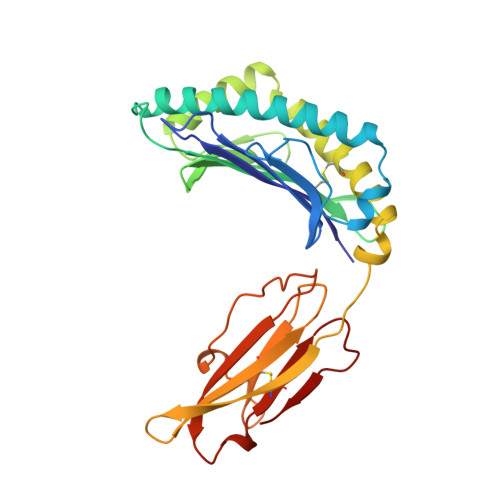
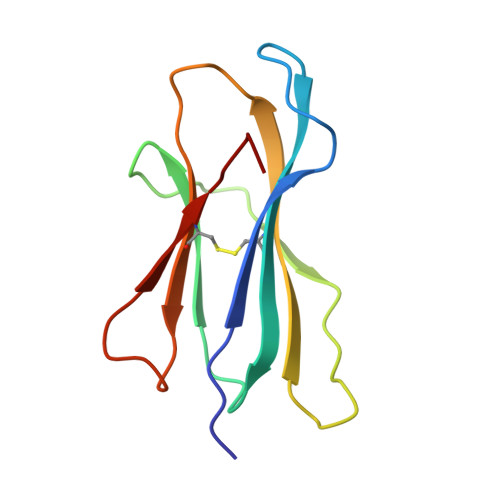
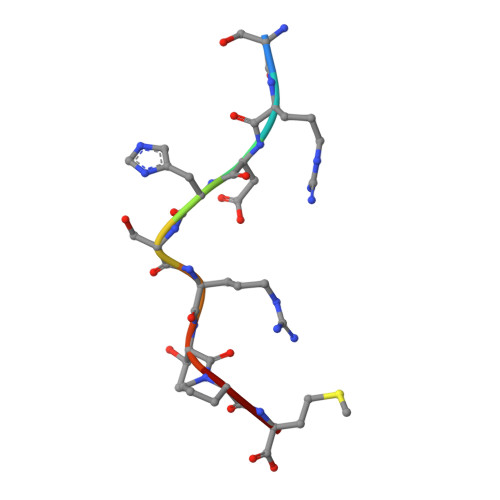
Crystal structure of an H-2Kb-ovalbumin peptide complex reveals the interplay of primary and secondary anchor positions in the major histocompatibility complex binding groove.
Fremont, D.H., Stura, E.A., Matsumura, M., Peterson, P.A., Wilson, I.A.(1995) Proc Natl Acad Sci U S A 92: 2479-2483
- PubMed: 7708669
- DOI: https://doi.org/10.1073/pnas.92.7.2479
- Primary Citation of Related Structures:
1VAC, 1VAD - PubMed Abstract:
Sequence analysis of peptides naturally presented by major histocompatibility complex (MHC) class I molecules has revealed allele-specific motifs in which the peptide length and the residues observed at certain positions are restricted. Nevertheless, peptides containing the standard motif often fail to bind with high affinity or form physiologically stable complexes. Here we present the crystal structure of a well-characterized antigenic peptide from ovalbumin [OVA-8, ovalbumin-(257-264), SIINFEKL] in complex with the murine MHC class I H-2Kb molecule at 2.5-A resolution. Hydrophobic peptide residues Ile-P2 and Phe-P5 are packed closely together into binding pockets B and C, suggesting that the interplay of peptide anchor (P5) and secondary anchor (P2) residues can couple the preferred sequences at these positions. Comparison with the crystal structures of H-2Kb in complex with peptides VSV-8 (RGYVYQGL) and SEV-9 (FAPGNYPAL), where a Tyr residue is used as the C pocket anchor, reveals that the conserved water molecule that binds into the B pocket and mediates hydrogen bonding from the buried anchor hydroxyl group could not be likewise positioned if the P2 side chain were of significant size. Based on this structural evidence, H-2Kb has at least two submotifs: one with Tyr at P5 (or P6 for nonamer peptides) and a small residue at P2 (i.e., Ala or Gly) and another with Phe at P5 and a medium-sized hydrophobic residue at P2 (i.e., Ile). Deciphering of these secondary submotifs from both crystallographic and immunological studies of MHC peptide binding should increase the accuracy of T-cell epitope prediction.
- Department of Molecular Biology, Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: