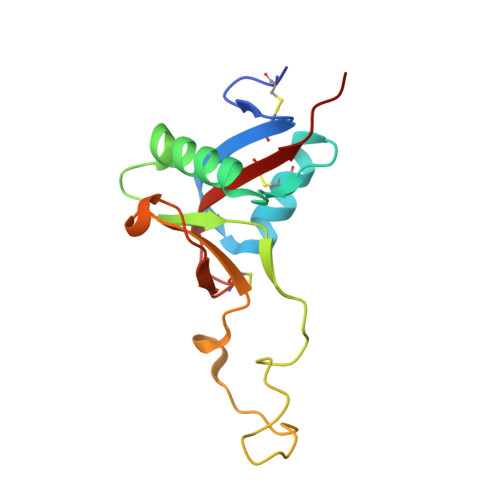
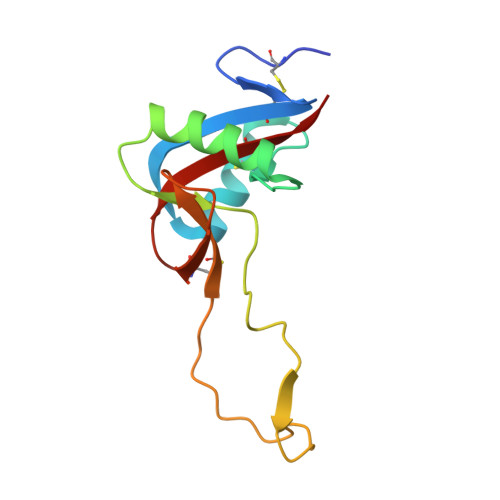
Structure of the Snake-Venom Toxin Convulxin
Batuwangala, T., Leduc, M., Gibbins, J.M., Bon, C., Jones, E.Y.(2004) Acta Crystallogr D Biol Crystallogr 60: 46
- PubMed: 14684891
- DOI: https://doi.org/10.1107/s0907444903021620
- Primary Citation of Related Structures:
1UOS - PubMed Abstract:
Snake venoms contain a number of proteins that interact with components of the haemostatic system that promote or inhibit events leading to blood-clot formation. The snake-venom protein convulxin (Cvx) binds glycoprotein (GP) VI, the platelet receptor for collagen, and triggers signal transduction. Here, the 2.7 A resolution crystal structure of Cvx is presented. In common with other members of this snake-venom protein family, Cvx is an alphabeta-heterodimer and conforms to the C-type lectin-fold topology. Comparison with other family members allows a set of Cvx residues that form a concave surface to be putatively implicated in GPVI binding. Unlike other family members, with the exception of flavocetin-A (FL-A), Cvx forms an (alphabeta)(4) tetramer. This oligomeric structure is consistent with Cvx clustering GPVI molecules on the surface of platelets and as a result promoting signal transduction activity. The Cvx structure and the location of the putative binding sites suggest a model for this multimeric signalling assembly.
- Cancer Research UK Receptor Structure Group, Division of Structural Biology, University of Oxford, The Henry Wellcome Building for Genomic Medicine, Roosevelt Drive, Headington, Oxford OX3 7BN, England.
Organizational Affiliation: