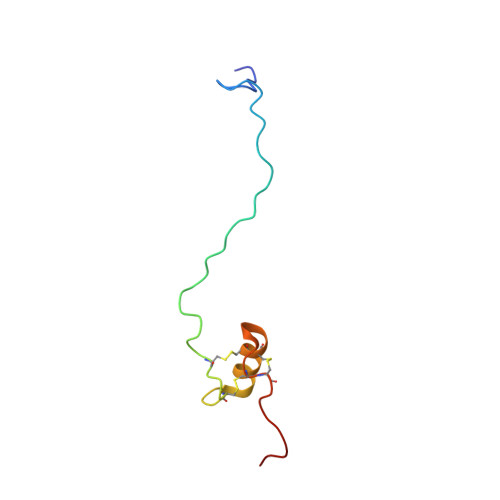
Solution structure of the recombinant penaeidin-3, a shrimp antimicrobial peptide
Yang, Y., Poncet, J., Garnier, J., Zatylny, C., Bachere, E., Aumelas, A.(2003) J Biol Chem 278: 36859-36867
- PubMed: 12842879
- DOI: https://doi.org/10.1074/jbc.M305450200
- Primary Citation of Related Structures:
1UEO - PubMed Abstract:
Penaeidins are a family of antimicrobial peptides of 47-63 residues isolated from several species of shrimp. These peptides display a proline-rich domain (N-terminal part) and a cysteine-rich domain (C-terminal part) stabilized by three conserved disulfide bonds whose arrangement has not yet been characterized. The recombinant penaeidin-3a of Litopenaeus vannamei (63 residues) and its [T8A]-Pen-3a analogue were produced in Saccharomyces cerevisiae and showed similar antimicrobial activity. The solution structure of the [T8A]-Pen-3a analogue was determined by using two-dimensional 1H NMR and simulated annealing calculations. The proline-rich domain, spanning residues 1-28 was found to be unconstrained. In contrast, the cysteine-rich domain, spanning residues 29-58, displays a well defined structure, which consists of an amphipathic helix (41-50) linked to the upstream and the downstream coils by two disulfide bonds (Cys32-Cys47 and Cys48-Cys55). These two coils are in turn linked together by the third disulfide bond (Cys36-Cys54). Such a disulfide bond packing, which is in agreement with the analysis of trypsin digests by ESI-MS, contributes to the highly hydrophobic core. Side chains of Arg45 and Arg50, which belong to the helix, and side chains of Arg37 and Arg53, which belong to the upstream and the downstream coils, are located in two opposite parts of this globular and compact structure. The environment of these positively charged residues, either by hydrophobic clusters at the surface of the cysteine-rich domain or by sequential hydrophobic residues in the unconstrained proline-rich domain, gives rise to the amphipathic character required for antimicrobial peptides. We hypothesize that the antimicrobial activity of penaeidins can be explained by a cooperative effect between the proline-rich and cysteine-rich features simultaneously present in their sequences.
Organizational Affiliation:
Centre de Biochimie Structurale, CNRS UMR 5048, INSERM U414, Université Montpellier 1, Faculté de Pharmacie, 15 avenue Charles Flahault, 34093 Montpellier Cedex 5, France.