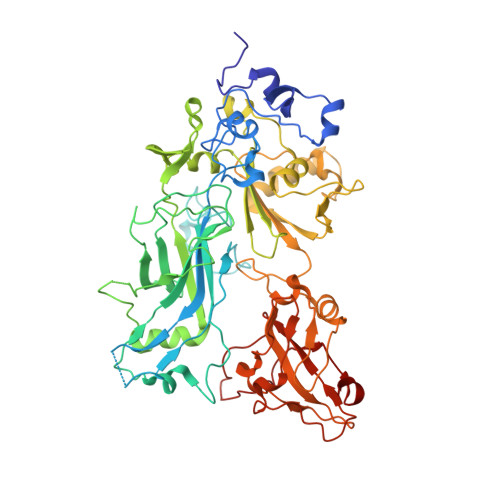
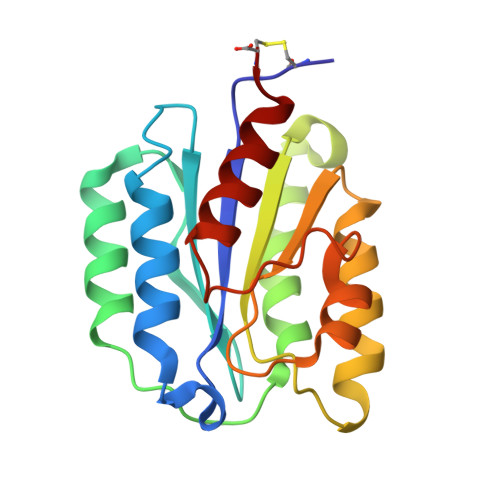
Structure of heptameric protective antigen bound to an anthrax toxin receptor: A role for receptor in pH-dependent pore formation
Lacy, D.B., Wigelsworth, D.J., Melnyk, R.A., Harrison, S.C., Collier, R.J.(2004) Proc Natl Acad Sci U S A 101: 13147-13151
- PubMed: 15326297
- DOI: https://doi.org/10.1073/pnas.0405405101
- Primary Citation of Related Structures:
1TZN, 1TZO - PubMed Abstract:
After binding to cellular receptors and proteolytic activation, the protective antigen component of anthrax toxin forms a heptameric prepore. The prepore later undergoes pH-dependent conversion to a pore, mediating translocation of the edema and lethal factors to the cytosol. We describe structures of the prepore (3.6 A) and a prepore:receptor complex (4.3 A) that reveal the location of pore-forming loops and an unexpected interaction of the receptor with the pore-forming domain. Lower pH is required for prepore-to-pore conversion in the presence of the receptor, indicating that this interaction regulates pH-dependent pore formation. We present an example of a receptor negatively regulating pH-dependent membrane insertion.
- Department of Microbiology and Molecular Genetics, Harvard Medical School, 200 Longwood Avenue, Boston, MA 02115, USA.
Organizational Affiliation: