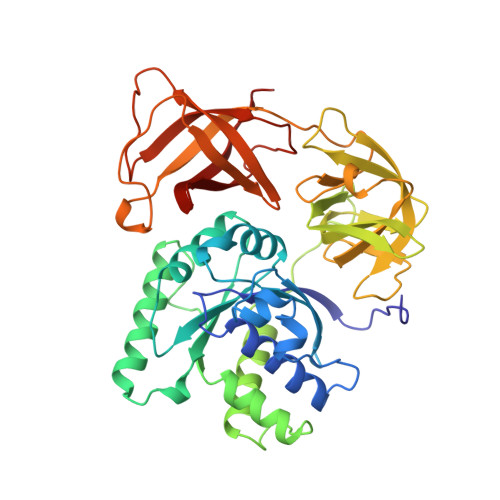
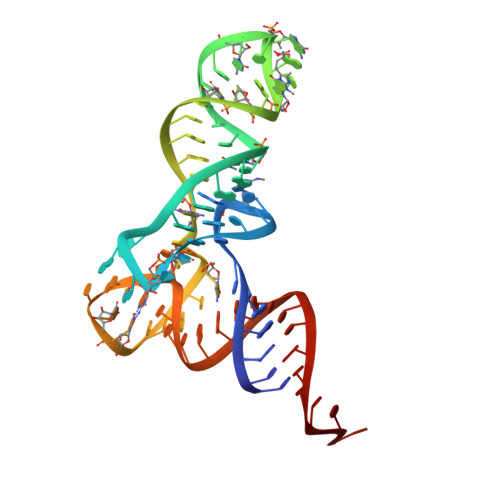
Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog.
Nissen, P., Kjeldgaard, M., Thirup, S., Polekhina, G., Reshetnikova, L., Clark, B.F., Nyborg, J.(1995) Science 270: 1464-1472
- PubMed: 7491491
- DOI: https://doi.org/10.1126/science.270.5241.1464
- Primary Citation of Related Structures:
1TTT - PubMed Abstract:
The structure of the ternary complex consisting of yeast phenylalanyl-transfer RNA (Phe-tRNAPhe), Thermus aquaticus elongation factor Tu (EF-Tu), and the guanosine triphosphate (GTP) analog GDPNP was determined by x-ray crystallography at 2.7 angstrom resolution. The ternary complex participates in placing the amino acids in their correct order when messenger RNA is translated into a protein sequence on the ribosome. The EF-Tu-GDPNP component binds to one side of the acceptor helix of Phe-tRNAPhe involving all three domains of EF-Tu. Binding sites for the phenylalanylated CCA end and the phosphorylated 5' end are located at domain interfaces, whereas the T stem interacts with the surface of the beta-barrel domain 3. The binding involves many conserved residues in EF-Tu. The overall shape of the ternary complex is similar to that of the translocation factor, EF-G-GDP, and this suggests a novel mechanism involving "molecular mimicry" in the translational apparatus.
- Department of Biostructural Chemistry, Institute of Chemistry, Aarhus University, Denmark.
Organizational Affiliation: