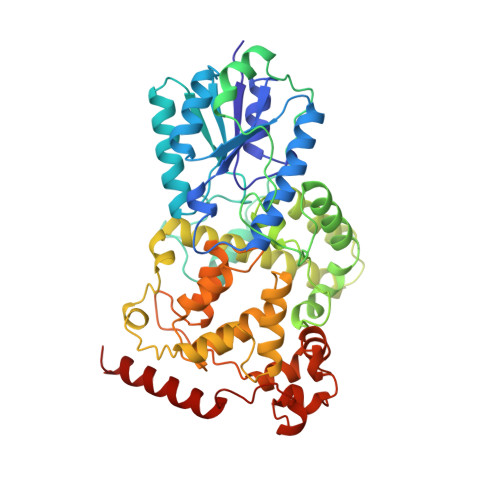
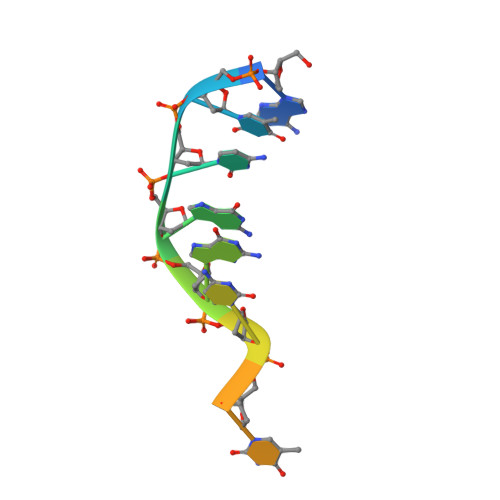
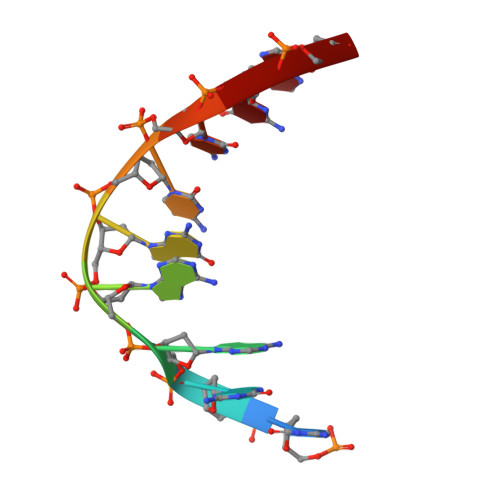
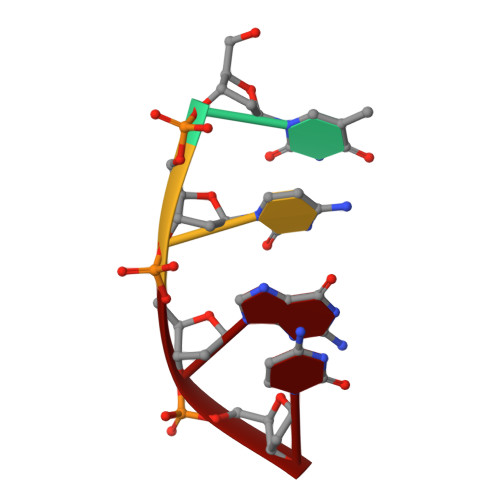
Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair
Mees, A., Klar, T., Gnau, P., Hennecke, U., Eker, A.P.M., Carell, T., Essen, L.-O.(2004) Science 306: 1789-1793
- PubMed: 15576622
- DOI: https://doi.org/10.1126/science.1101598
- Primary Citation of Related Structures:
1TEZ - PubMed Abstract:
DNA photolyases use light energy to repair DNA that comprises ultraviolet-induced lesions such as the cis-syn cyclobutane pyrimidine dimers (CPDs). Here we report the crystal structure of a DNA photolyase bound to duplex DNA that is bent by 50 degrees and comprises a synthetic CPD lesion. This CPD lesion is flipped into the active site and split there into two thymines by synchrotron radiation at 100 K. Although photolyases catalyze blue light-driven CPD cleavage only above 200 K, this structure apparently mimics a structural substate during light-driven DNA repair in which back-flipping of the thymines into duplex DNA has not yet taken place.
- Department of Chemistry and Biochemistry, Butenandt-Strasse 5-13, Ludwig Maximilians University, D-81377 Munich, Germany.
Organizational Affiliation: