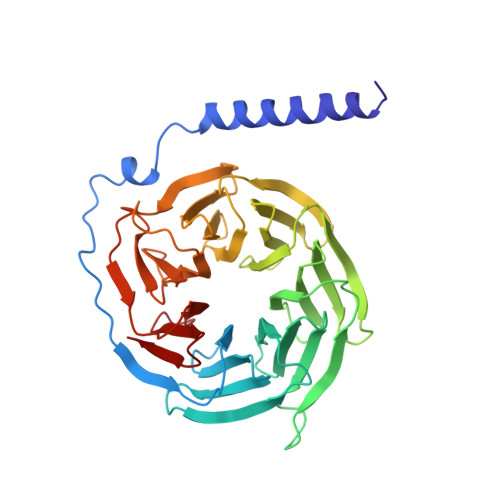
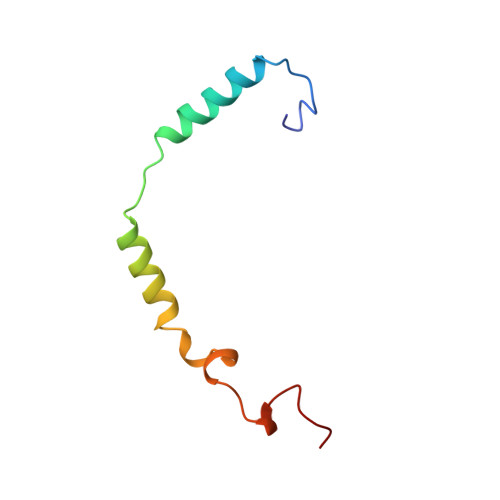
Crystal structure of a G-protein beta gamma dimer at 2.1A resolution.
Sondek, J., Bohm, A., Lambright, D.G., Hamm, H.E., Sigler, P.B.(1996) Nature 379: 369-374
- PubMed: 8552196
- DOI: https://doi.org/10.1038/379369a0
- Primary Citation of Related Structures:
1TBG - PubMed Abstract:
Many signalling cascades use seven-helical transmembrane receptors coupled to heterotrimeric G proteins (G alpha beta gamma) to convert extracellular signals into intracellular responses. Upon nucleotide exchange catalysed by activated receptors, heterotrimers dissociate into GTP-bound G alpha subunits and G beta gamma dimers, either of which can modulate many downstream effectors. Here we use multiwavelength anomalous diffraction data to solve the crystal structure of the beta gamma dimer of the G protein transducin. The beta-subunit is primarily a seven-bladed beta-propeller that is partially encircled by an extended gamma-subunit. The beta-propeller, which contains seven structurally similar WD repeats, defines the stereochemistry of the WD repeat and the probable architecture of all WD-repeat-containing domains. The structure details interactions between G protein beta- and gamma-subunits and highlights regions implicated in effector modulation for the conserved family of G protein beta gamma dimers.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06510, USA.
Organizational Affiliation: