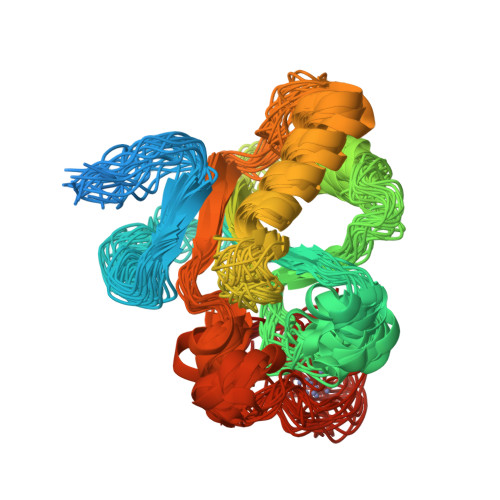
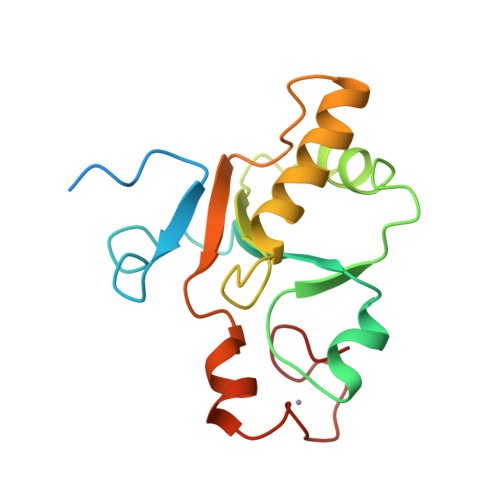
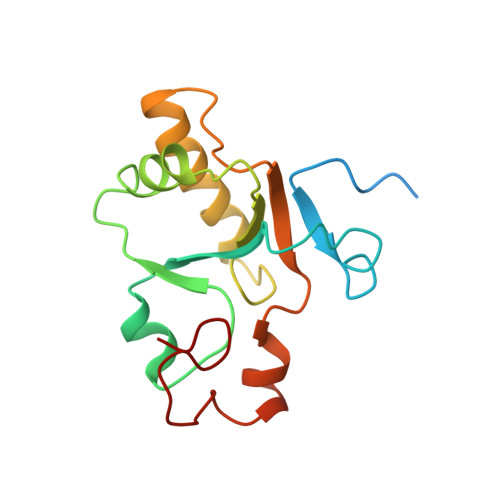
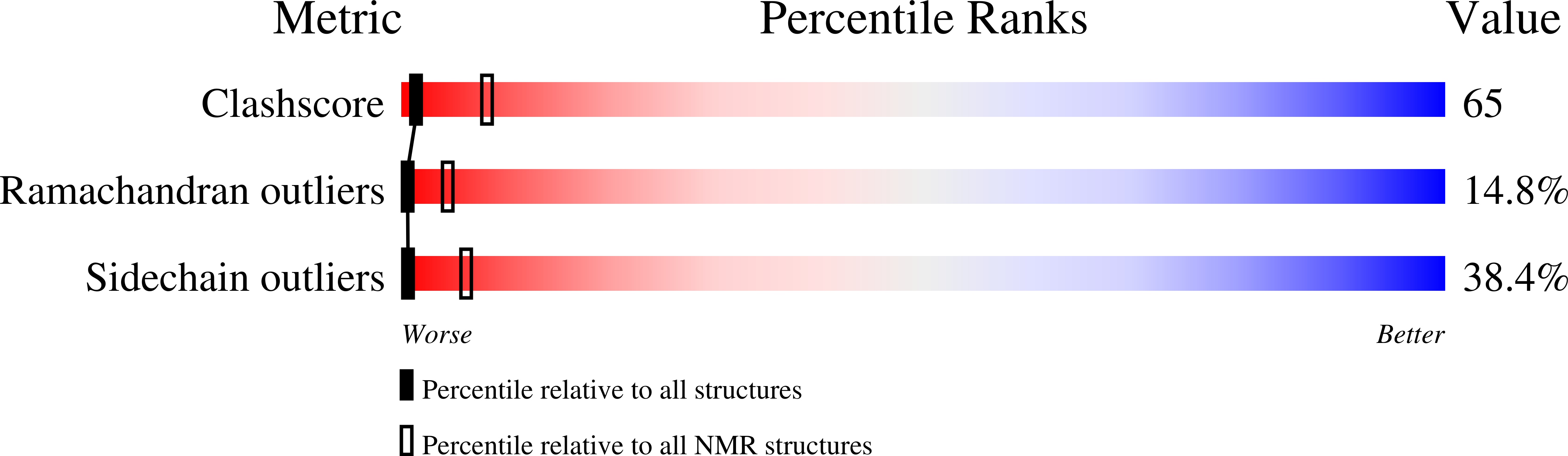A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana.
Landrieu, I., Da Costa, M., De Veylder, L., Dewitte, F., Vandepoele, K., Hassan, S., Wieruszeski, J.M., Faure, J.D., Van Montagu, M., Inze, D., Lippens, G.(2004) Proc Natl Acad Sci U S A 101: 13380-13385
- PubMed: 15329414
- DOI: https://doi.org/10.1073/pnas.0405248101
- Primary Citation of Related Structures:
1T3K - PubMed Abstract:
The dual-specificity CDC25 phosphatases are critical positive regulators of cyclin-dependent kinases (CDKs). Even though an antagonistic Arabidopsis thaliana WEE1 kinase has been cloned and tyrosine phosphorylation of its CDKs has been demonstrated, no valid candidate for a CDC25 protein has been reported in higher plants. We identify a CDC25-related protein (Arath;CDC25) of A. thaliana, constituted by a sole catalytic domain. The protein has a tyrosine-phosphatase activity and stimulates the kinase activity of Arabidopsis CDKs. Its tertiary structure was obtained by NMR spectroscopy and confirms that Arath;CDC25 belongs structurally to the classical CDC25 superfamily with a central five-stranded beta-sheet surrounded by helices. A particular feature of the protein, however, is the presence of an additional zinc-binding loop in the C-terminal part. NMR mapping studies revealed the interaction with phosphorylated peptidic models derived from the conserved CDK loop containing the phosphothreonine-14 and phosphotyrosine-15. We conclude that despite sequence divergence, Arath;CDC25 is structurally and functionally an isoform of the CDC25 superfamily, which is conserved in yeast and in plants, including Arabidopsis and rice.
Organizational Affiliation:
Unité Mixte de Recherche 8525 Centre National de la Recherche Scientifique-Lille2, Institut de Biologie de Lille/Pasteur Institute of Lille, 59019 Lille Cedex, France. isabelle.landrieu@ibl.fr