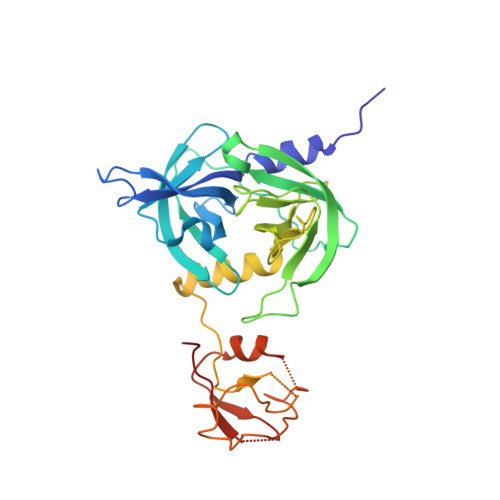
Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Wilken, C., Kitzing, K., Kurzbauer, R., Ehrmann, M., Clausen, T.(2004) Cell 117: 483-494
- PubMed: 15137941
- DOI: https://doi.org/10.1016/s0092-8674(04)00454-4
- Primary Citation of Related Structures:
1SOT, 1SOZ, 1VCW - PubMed Abstract:
Gram-negative bacteria respond to misfolded proteins in the cell envelope with the sigmaE-driven expression of periplasmic proteases/chaperones. Activation of sigmaE is controlled by a proteolytic cascade that is initiated by the DegS protease. DegS senses misfolded protein in the periplasm, undergoes autoactivation, and cleaves the antisigma factor RseA. Here, we present the crystal structures of three distinct states of DegS from E. coli. DegS alone exists in a catalytically inactive form. Binding of stress-signaling peptides to its PDZ domain induces a series of conformational changes that activates protease function. Backsoaking of crystals containing the DegS-activator complex revealed the presence of an active/inactive hybrid structure and demonstrated the reversibility of activation. Taken together, the structural data illustrate in molecular detail how DegS acts as a periplasmic stress sensor. Our results suggest a novel regulatory role for PDZ domains and unveil a novel mechanism of reversible protease activation.
- Institute for Molecular Pathology (IMP), Dr. Bohrgasse 7, A-1030 Vienna, Austria.
Organizational Affiliation: