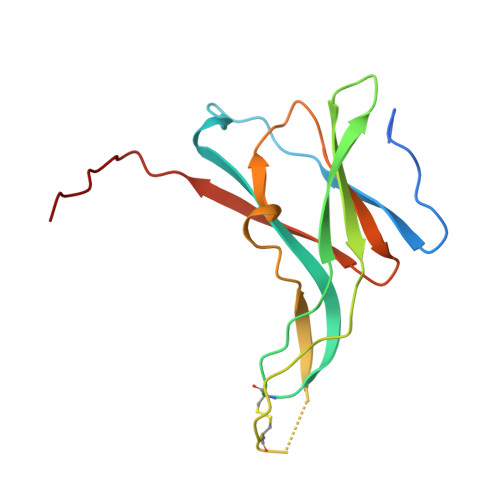
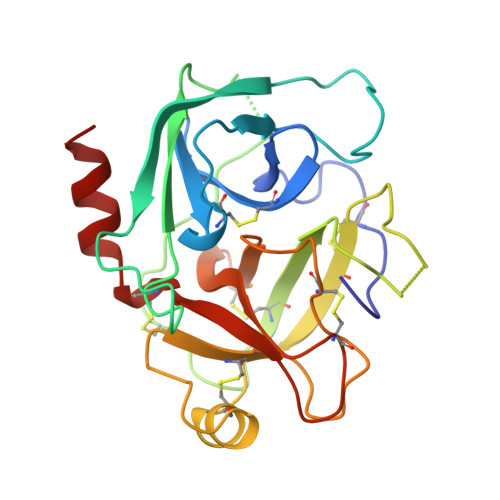
X-ray structures of a designed binding site in trypsin show metal-dependent geometry.
Brinen, L.S., Willett, W.S., Craik, C.S., Fletterick, R.J.(1996) Biochemistry 35: 5999-6009
- PubMed: 8634241
- DOI: https://doi.org/10.1021/bi9530200
- Primary Citation of Related Structures:
1SLU, 1SLV, 1SLW, 1SLX - PubMed Abstract:
The three-dimensional structures of complexes of trypsin N143H, E151H bound to ecotin A86H are determined at 2.0 A resolution via X-ray crystallography in the absence and presence of the transition metals Zn2+, Ni2+, and Cu2+. The binding site for these transition metals was constructed by substitution of key amino acids with histidine at the trypsin-ecotin interface in the S2'/P2' pocket. Three histidine side chains, two on trypsin at positions 143 and 151 and one on ecotin at position 86, anchor the metals and provide extended catalytic recognition for substrates with His in the P2' pocket. Comparisons of the three-dimensional structures show the different geometries that result upon the binding of metal in the engineered tridentate site and suggest a structural basis for the kinetics of the metal-regulated catalysis. Of the three metals, the binding of zinc results in the most favorable binding geometry, not dissimilar to those observed in naturally occurring zinc binding proteins.
- Department of Biochemistry and Biophysics, University of California at San Francisco 94143, USA.
Organizational Affiliation: