Solution Structure of a Methionine-Rich 2S Albumin from Sunflower Seeds: Relationship to Its Allergenic and Emulsifying Properties.
Pantoja-Uceda, D., Shewry, P.R., Bruix, M., Tatham, A.S., Santoro, J., Rico, M.(2004) Biochemistry 43: 6976-6986
- PubMed: 15170335
- DOI: https://doi.org/10.1021/bi0496900
- Primary Citation of Related Structures:
1S6D - PubMed Abstract:
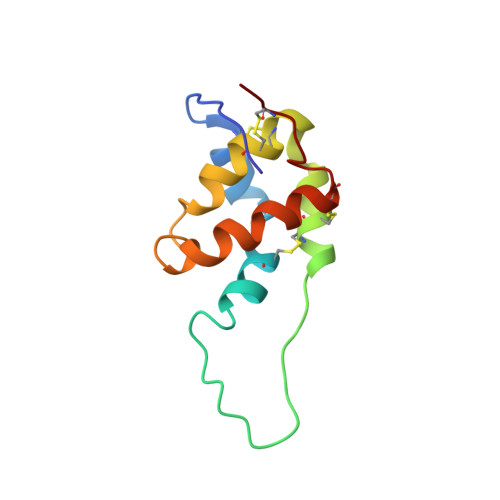
The three-dimensional structure in aqueous solution of SFA-8, a 2S albumin 103-residue protein from seeds of sunflower (Helianthus anuus L.), has been determined by NMR methods. An almost complete (1)H resonance assignment was accomplished from analysis of two-dimensional (2D) COSY and 2D TOCSY spectra, and the structure was computed by using restrained molecular dynamics on the basis of 1393 upper limit distance constraints derived from NOE cross-correlation intensities measured in 2D NOESY spectra. In contrast with most other 2S albumins, SFA-8 consists of a single polypeptide chain without any cleavage in the segment of residues 30-46. The computed structures exhibited an rmsd radius of 0.52 A for the backbone structural core (residues 11-30 and 46-101) and 1.01 A for the side chain heavy atoms. The resulting structure consists of five amphipathic helices arranged in a right-handed superhelix, a folding motif first observed in nonspecific lipid transfer (nsLTP) proteins, and common to other 2S albumins. In contrast to nsLTP proteins, neither SFA-8 nor RicC3 (a 2S albumin from castor bean) has an internal cavity that is able to host a lipid molecule, which results from an exchange in the pairing of disulfide bridges in the CXC segment. Both 2S albumins and nonspecific lipid transfer proteins belong to the prolamin superfamily, which includes a number of important food allergens. Differences in the extension and solvent exposition of the so-called "hypervariable loop" (which connects helices III and IV) in SFA-8 and RicC3 may be responsible for the different allergenic properties of the two proteins. SFA-8 has been shown to form highly stable emulsions with oil/water mixtures. We propose that these properties may be determined partly by a hydrophobic patch at the surface of the protein which consists of five methionines that partially hide the Trp76 residue. The flexibility of the loop which contains Trp76 and the hydrophobicity of the whole environment may favor a conformational change, by which the Trp76 side chain may become inserted into the oil phase.
- Instituto de Química Física Rocasolano, CSIC, Serrano 119, Madrid 28006, Spain.
Organizational Affiliation: