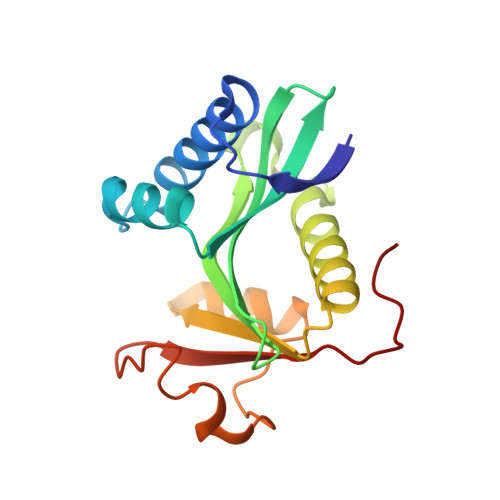
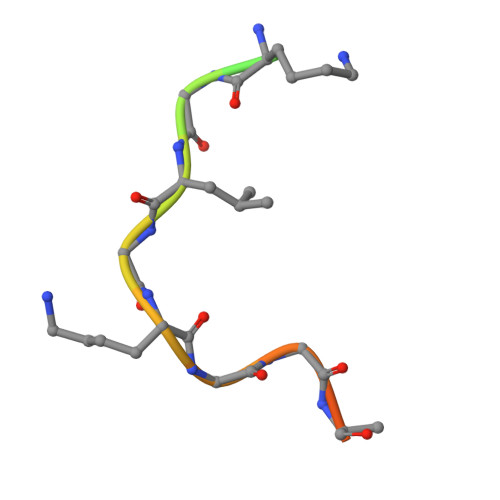
Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase
Clements, A., Poux, A.N., Lo, S., Pillus, L., Berger, S.L., Marmorstein, R.(2003) Mol Cell 12: 461-473
- PubMed: 14536085
- DOI: https://doi.org/10.1016/s1097-2765(03)00288-0
- Primary Citation of Related Structures:
1PU9, 1PUA, 1Q2C - PubMed Abstract:
Distinct posttranslational modifications on histones occur in specific patterns to mediate certain chromosomal events. For example, on histone H3, phosphorylation at Ser10 can enhance GCN5-mediated Lys14 acetylation to promote transcription. To gain insight into the mechanism underlying this synergism, we determined the structure of Tetrahymena GCN5 (tGCN5) and coenzyme A (CoA) bound to unmodified and Ser10-phosphorylated 19 residue histone H3 peptides (H3p19 and H3p19Pi, respectively). The tGCN5/CoA/H3p19 structure reveals that a 12 amino acid core sequence mediates extensive contacts with the protein, providing the structural basis for substrate specificity by the GCN5/PCAF family of histone acetyltransferases. Comparison with the tGCN5/CoA/H3p19Pi structure reveals that phospho-Ser10 and Thr11 mediate significant histone-protein interactions, and nucleate additional interactions distal to the phosphorylation site. Functional studies show that histone H3 Thr11 is necessary for optimal transcription at yGcn5-dependent promoters requiring Ser10 phosphorylation. Together, these studies reveal how one histone modification can modulate another to affect distinct transcriptional signals.
- The Wistar Institute, Philadelphia, PA 19104, USA.
Organizational Affiliation: