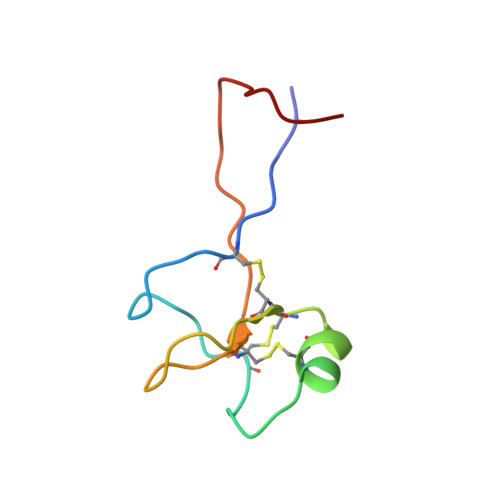
High-resolution solution structure of human pNR-2/pS2: a single trefoil motif protein.
Polshakov, V.I., Williams, M.A., Gargaro, A.R., Frenkiel, T.A., Westley, B.R., Chadwick, M.P., May, F.E., Feeney, J.(1997) J Mol Biology 267: 418-432
- PubMed: 9096235
- DOI: https://doi.org/10.1006/jmbi.1997.0896
- Primary Citation of Related Structures:
1PS2 - PubMed Abstract:
pNR-2/pS2 is a 60 residue extracellular protein, which was originally discovered in human breast cancer cells, and subsequently found in other tumours and normal gastric epithelial cells. We have determined the three-dimensional solution structure of a C58S mutant of human pNR-2/pS2 using 639 distance and 137 torsion angle constraints obtained from analysis of multidimensional NMR spectra. A series of simulated annealing calculations resulted in the unambiguous determination of the protein's disulphide bonding pattern and produced a family of 19 structures consistent with the constraints. The peptide contains a single "trefoil" sequence motif, a region of about 40 residues with a characteristic sequence pattern, which has been found, either singly or as a repeat, in about a dozen extracellular proteins. The trefoil domain contains three disulphide bonds, whose 1-5, 2-4 and 3-6 cysteine pairings form the structure into three closely packed loops with only a small amount of secondary structure, which consists of a short alpha-helix packed against a two-stranded antiparallel beta-sheet. The structure of the domain is very similar to those of the two trefoil domains that occur in porcine spasmolytic polypeptide (PSP), the only member of the trefoil family whose three-dimensional structure has been previously determined. Outside the trefoil domain, which forms the compact "head" of the molecule, the N and C-terminal strands are closely associated, forming an extended "tail", which has some beta-sheet character for part of its length and which becomes more disordered towards the termini as indicated by (15)N{(1)H} NOEs. We have considered the structural implications of the possible formation of a native C58-C58 disulphide-bonded homodimer. Comparison of the surface features of pNR-2/pS2 and PSP, and consideration of the sequences of the other human trefoil domains in the light of these structures, illuminates the possible role of specific residues in ligand/receptor binding.
- Laboratory of Physical Methods, Chemical-Pharmaceutical Research Institute, Moscow, Russia.
Organizational Affiliation: