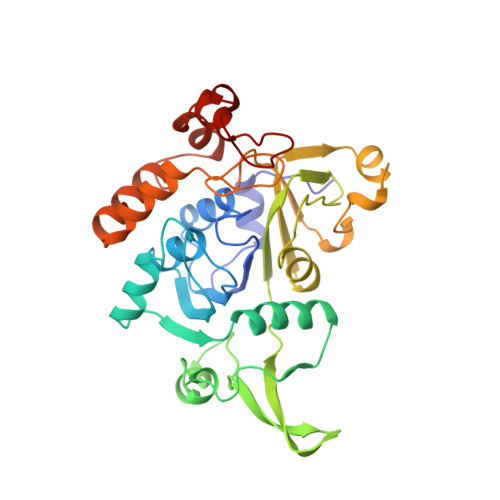
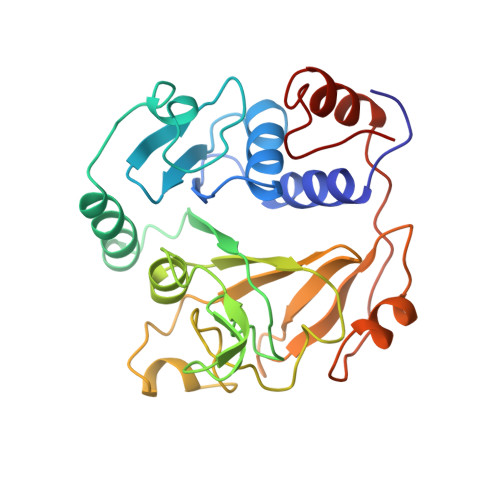
Glutaconate CoA-transferase from Acidaminococcus fermentans: the crystal structure reveals homology with other CoA-transferases.
Jacob, U., Mack, M., Clausen, T., Huber, R., Buckel, W., Messerschmidt, A.(1997) Structure 5: 415-426
- PubMed: 9083111
- DOI: https://doi.org/10.1016/s0969-2126(97)00198-6
- Primary Citation of Related Structures:
1POI - PubMed Abstract:
Coenzyme A-transferases are a family of enzymes with a diverse substrate specificity and subunit composition. Members of this group of enzymes are found in anaerobic fermenting bacteria, aerobic bacteria and in the mitochondria of humans and other mammals, but so far none have been crystallized. A defect in the human gene encoding succinyl-CoA: 3-oxoacid CoA-transferase causes a metabolic disease which leads to severe ketoacidosis, thus reflecting the importance of this family of enzymes. All CoA-transferases share a common mechanism in which the CoA moiety is transferred from a donor (e.g. acetyl CoA) to an acceptor, (R)-2-hydroxyglutarate, whereby acetate is formed. The transfer has been described by a ping-pong mechanism in which CoA is bound to the active-site residue of the enzyme as a covalent thiol ester intermediate. We describe here the crystal structure of glutaconate CoA-transferase (GCT) from the strictly anaerobic bacterium Acidaminococcus fermentans. This enzyme activates (R)-2-hydroxyglutarate to (R)-2-hydroxyglutaryl-CoA in the pathway of glutamate fermentation. We initiated this project to gain further insight into the function of this enzyme and the structural basis for the characteristics of CoA-transferases. The crystal structure of GCT was solved by multiple isomorphous replacement to 2.55 A resolution. The enzyme is a heterooctamer and its overall arrangement of subunits can be regarded as an (AB)4tetramer obeying 222 symmetry. Both subunits A and B belong to the open alpha/beta-protein class and can be described as a four-layered alpha/alpha/beta/alpha type with a novel composition and connectivity of the secondary structure elements. The core of subunit A consists of seven alpha/beta repeats resulting in an all parallel central beta sheet, against which helices pack from both sides. In contrast, the centre of subunit B is formed by a ninefold mixed beta sheet. In both subunits the helical C terminus is folded back onto the N-terminal domain to form the third layer of helices. The active site of GCT is located at the interface of subunits A and B and is formed by loops of both subunits. The funnel-shaped opening to the active site has a depth and diameter of about 20 A with the catalytic residue, Glu54 of subunit B, at the bottom. The active-site glutamate residue is stabilized by hydrogen bonds. Despite very low amino acid sequence similarity, subunits A and B reveal a similar overall fold. Large parts of their structures can be spatially superimposed, suggesting that both subunits have evolved from a common ancestor.
- Max Planck Institut für Biochemie, Abteilung Strukturforschung, Am Klopferspitz 18a, D-82152, Martinsried, Germany. ujacob@libelle.biochem.mpg.de
Organizational Affiliation: