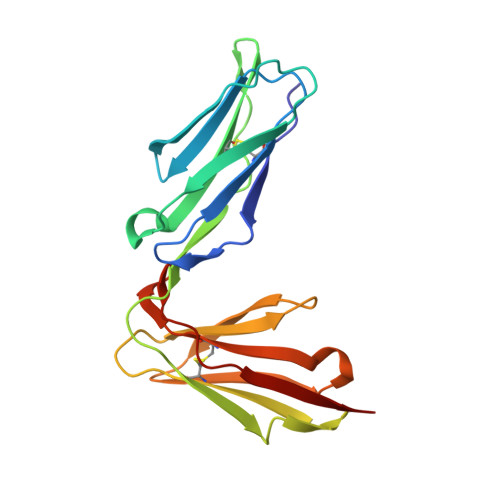
Structure of the Human Nk Cell Triggering Receptor Nkp46 Ectodomain
Ponassi, M., Cantoni, C., Biassoni, R., Conte, R., Spallarossa, A., Pesce, A., Moretta, A., Moretta, L., Bolognesi, M., Bordo, D.(2003) Biochem Biophys Res Commun 309: 317
- PubMed: 12951052
- DOI: https://doi.org/10.1016/j.bbrc.2003.08.007
- Primary Citation of Related Structures:
1OLL - PubMed Abstract:
NKp46, a natural killer (NK) cell-specific receptor, has been recently identified as one of the triggering receptors involved in NK cell activation mediated by non-HLA class I ligands. The structure of the NKp46 extracellular receptor region, here reported, consists of two Ig-like domains assembled similarly to leukocyte immunoglobulin-like receptors (LIRs) and killer inhibitory receptors (KIRs). The extensive NKp46 residue substitutions at sites structurally related to those mediating interaction with HLA antigens in LIRs and KIRs indicate that NKp46 recognition processes in vivo should involve non-HLA ligands. NKP46 is shown to stem from an ancestral KIR/LIR family. However, the absence of close paralogues, such as those found for LIR and KIR, indicates that NKp46 is the unique member of a distinct Ig-like subfamily and suggests a specific role, which appears to be maintained across primates and rodents.
- Istituto Nazionale Ricerca sul Cancro, Largo R. Benzi 10, 16132 Genoa, Italy.
Organizational Affiliation: