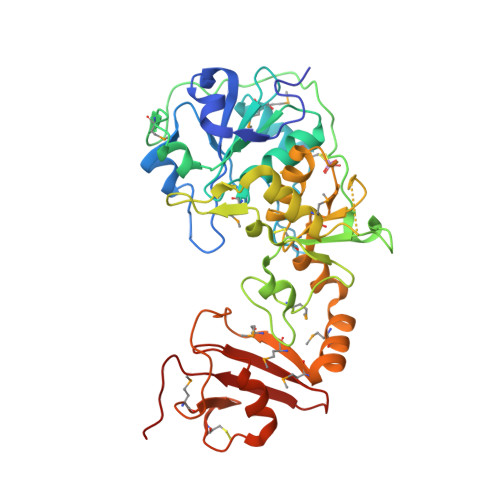
The Crystal Structure of Leishmania Major 3-Mercaptopyruvate Sulfurtransferase: A Three-Domain Architecture with a Serine Protease-Like Triad at the Active Site
Alphey, M.S., Williams, R.A.M., Mottram, J.C., Coombs, G.H., Hunter, W.N.(2003) J Biological Chem 278: 48219
- PubMed: 12952945
- DOI: https://doi.org/10.1074/jbc.M307187200
- Primary Citation of Related Structures:
1OKG - PubMed Abstract:
Leishmania major 3-mercaptopyruvate sulfurtransferase is a crescent-shaped molecule comprising three domains. The N-terminal and central domains are similar to the thiosulfate sulfurtransferase rhodanese and create the active site containing a persulfurated catalytic cysteine (Cys-253) and an inhibitory sulfite coordinated by Arg-74 and Arg-185. A serine protease-like triad, comprising Asp-61, His-75, and Ser-255, is near Cys-253 and represents a conserved feature that distinguishes 3-mercaptopyruvate sulfurtransferases from thiosulfate sulfurtransferases. During catalysis, Ser-255 may polarize the carbonyl group of 3-mercaptopyruvate to assist thiophilic attack, whereas Arg-74 and Arg-185 bind the carboxylate group. The enzyme hydrolyzes benzoyl-Arg-p-nitroanilide, an activity that is sensitive to the presence of the serine protease inhibitor N alpha-p-tosyl-L-lysine chloromethyl ketone, which also lowers 3-mercaptopyruvate sulfurtransferase activity, presumably by interference with the contribution of Ser-255. The L. major 3-mercaptopyruvate sulfurtransferase is unusual with an 80-amino acid C-terminal domain, bearing remarkable structural similarity to the FK506-binding protein class of peptidylprolyl cis/trans-isomerase. This domain may be involved in mediating protein folding and sulfurtransferase-protein interactions.
- Division of Biological Chemistry and Molecular Microbiology, School of Life Sciences, University of Dundee, Dundee DD1 5EH, United Kingdom.
Organizational Affiliation: