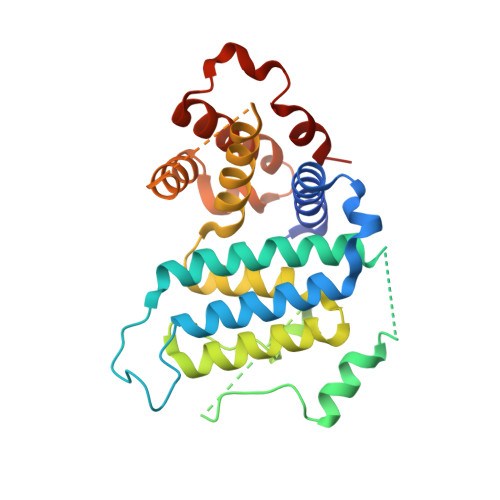
The Crystal Structure of Trypanosoma Cruzi Dutpase Reveals a Novel Dutp/Dudp Binding Fold
Harkiolaki, M., Dodson, E.J., Bernier-Villamor, V., Turkenburg, J.P., Gonzalez-Pacanowska, D., Wilson, K.S.(2004) Structure 12: 41
- PubMed: 14725764
- DOI: https://doi.org/10.1016/j.str.2003.11.016
- Primary Citation of Related Structures:
1OGK, 1OGL - PubMed Abstract:
dUTPase is an essential enzyme involved with nucleotide metabolism and replication. We report here the X-ray structure of Trypanosoma cruzi dUTPase in its native conformation and as a complex with dUDP. These reveal a novel protein fold that displays no structural similarities to previously described dUTPases. The molecular unit is a dimer with two active sites. Nucleotide binding promotes extensive structural rearrangements, secondary structure remodeling, and rigid body displacements of 20 A or more, which effectively bury the substrate within the enzyme core for the purpose of hydrolysis. The molecular complex is a trapped enzyme-substrate arrangement which clearly demonstrates structure-induced specificity and catalytic potential. This enzyme is a novel dUTPase and therefore a potential drug target in the treatment of Chagas' disease.
- Structural Biology Laboratory, Department of Chemistry, University of York, Heslington, York YO10 5YW, UK.
Organizational Affiliation: