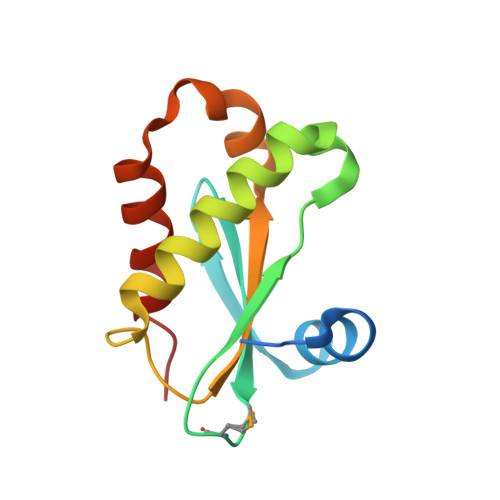
High-resolution structure of RNase P protein from Thermotoga maritima.
Kazantsev, A.V., Krivenko, A.A., Harrington, D.J., Carter, R.J., Holbrook, S.R., Adams, P.D., Pace, N.R.(2003) Proc Natl Acad Sci U S A 100: 7497-7502
- PubMed: 12799461
- DOI: https://doi.org/10.1073/pnas.0932597100
- Primary Citation of Related Structures:
1NZ0 - PubMed Abstract:
The structure of RNase P protein from the hyperthermophilic bacterium Thermotoga maritima was determined at 1.2-A resolution by using x-ray crystallography. This protein structure is from an ancestral-type RNase P and bears remarkable similarity to the recently determined structures of RNase P proteins from bacteria that have the distinct, Bacillus type of RNase P. These two types of protein span the extent of bacterial RNase P diversity, so the results generalize the structure of the bacterial RNase P protein. The broad phylogenetic conservation of structure and distribution of potential RNA-binding elements in the RNase P proteins indicate that all of these homologous proteins bind to their cognate RNAs primarily by interaction with the phylogenetically conserved core of the RNA. The protein is found to dimerize through an extensive, well-ordered interface. This dimerization may reflect a mechanism of thermal stability of the protein before assembly with the RNA moiety of the holoenzyme.
- Department of Molecular, Cellular, and Developmental Biology, University of Colorado, Boulder, CO 80309-0347, USA.
Organizational Affiliation: