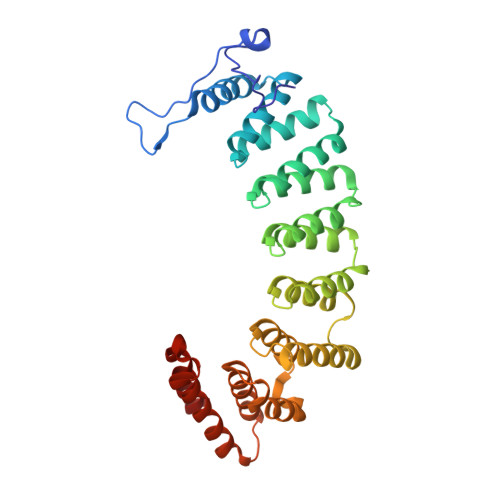
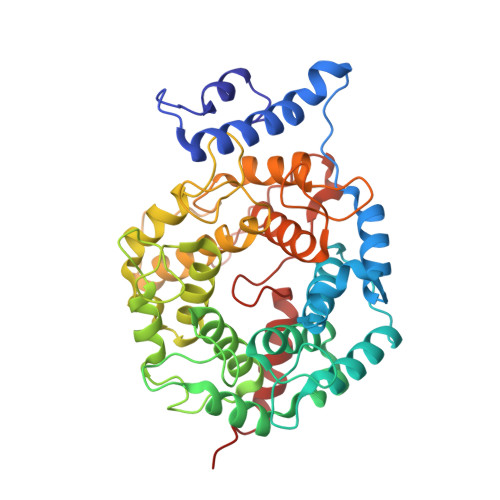
Novel and Selective Imidazole-containing Biphenyl Inhibitors of Protein Farnesyltransferase
Curtin, M.L., Florjancic, A.S., Cohen, J., Gu, W.-J., Frost, D.J., Muchmore, S.W., Sham, H.L.(2003) Bioorg Med Chem Lett 13: 1367-1371
- PubMed: 12657284
- DOI: https://doi.org/10.1016/s0960-894x(03)00096-9
- Primary Citation of Related Structures:
1NL4 - PubMed Abstract:
A series of imidazole-containing biphenyls was prepared and evaluated in vitro for inhibition of FTase and cellular Ras processing. Several of these analogues, such as 21, are potent inhibitors of FTase (<1nM), FTase/GGTase selective (>300-fold) and cellularly active (
- Department of Cancer Research, Global Pharmaceutical Research and Development, Abbott Laboratories, Abbott Park, IL 60064, USA. mike.curtin@abbott.com
Organizational Affiliation: