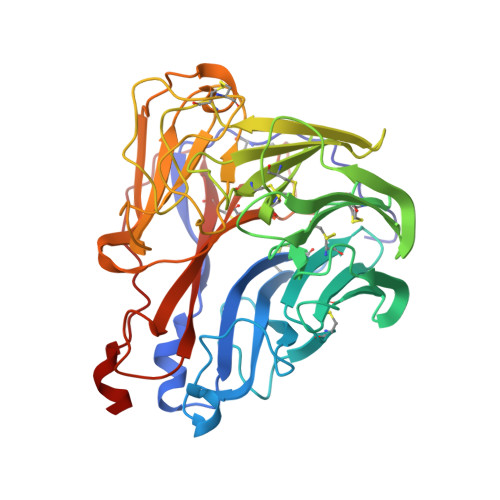
Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
Tulip, W.R., Varghese, J.N., Webster, R.G., Laver, W.G., Colman, P.M.(1992) J Mol Biology 227: 149-159
- PubMed: 1522584
- DOI: https://doi.org/10.1016/0022-2836(92)90688-g
- Primary Citation of Related Structures:
1NCB, 1NCC - PubMed Abstract:
The site on influenza virus N9 neuraminidase recognized by NC41 monoclonal antibody comprises 19 amino acid residues that are in direct contact with 17 residues on the antibody. Single sequence changes in some of the neuraminidase residues in the site markedly reduce antibody binding. However, two mutants have been found within the site, Ile368 to Arg and Asn329 to Asp selected by antibodies other than NC41, and these mutants bind NC41 antibody with only slightly reduced affinity. The three-dimensional structures of the two mutant N9-NC41 antibody complexes as derived from the wild-type complex are presented. Both structures show that some amino acid substitutions can be accommodated within an antigen-antibody interface by local structural rearrangements around the mutation site. In the Ile368 to Arg mutant complex, the side-chain of Arg368 is shifted by 2.9 A from its position in the uncomplexed mutant and a shift of 1.3 A in the position of the light chain residue HisL55 with respect to the wild-type complex is also observed. In the other mutant, the side-chain of Asp329 appears rotated by 150 degrees around C alpha-C beta with respect to the uncomplexed mutant, so that the carboxylate group is moved to the periphery of the antigen-antibody interface. The results provide a basis for understanding some of the potential structural effects of somatic hypermutation on antigen-antibody binding in those cases where the mutation in the antibody occurs at antigen-contacting residues, and demonstrate again the importance of structural context in evaluating the effect of amino acid substitutions on protein structure and function.
- CSIRO Division of Biomolecular Engineering, Parkville, Victoria, Australia.
Organizational Affiliation: