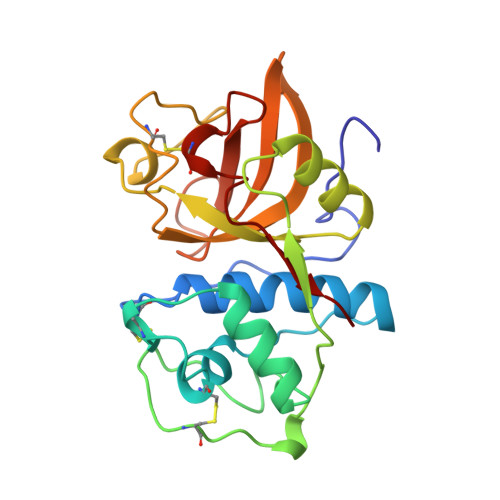
Crystal structure of human cathepsin K complexed with a potent inhibitor.
McGrath, M.E., Klaus, J.L., Barnes, M.G., Bromme, D.(1997) Nat Struct Biol 4: 105-109
- PubMed: 9033587
- DOI: https://doi.org/10.1038/nsb0297-105
- Primary Citation of Related Structures:
1MEM