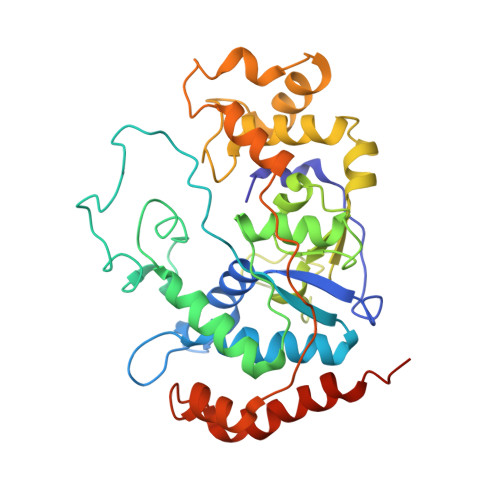
Molecular Structure and Novel DNA Binding Sites Located in Loops of Flap Endonuclease-1 from Pyrococcus horikoshii
Matsui, E., Musti, K.V., Abe, J., Yamazaki, K., Matsui, I., Harata, K.(2002) J Biological Chem 277: 37840-37847
- PubMed: 12147694
- DOI: https://doi.org/10.1074/jbc.M205235200
- Primary Citation of Related Structures:
1MC8 - PubMed Abstract:
The crystal structure of flap endonuclease-1 from Pyrococcus horikoshii (phFEN-1) was determined to a resolution of 3.1 A. The active cleft of the phFEN-1 molecule is formed with one large loop and four small loops. We examined the function of the conserved residues and positively charged clusters on these loops by kinetic analysis with 45 different mutants. Arg(40) and Arg(42) on small loop 1, a cluster Lys(193)-Lys(195) on small loop 2, and two sites, Arg(94) and Arg(118)-Lys(119), on the large loop were identified as binding sites. Lys(87) on the large loop may play significant roles in catalytic reaction. Furthermore, we successfully elucidated the function of the four DNA binding sites that form productive ES complexes specific for each endo- or exo-type hydrolysis, probably by bending the substrates. For the endo-activity, Arg(94) and Lys(193)-Lys(195) located at the top and bottom of the molecule were key determinants. For the exo-activity, all four sites were needed, but Arg(118)-Lys(119) was dominant. The major binding sites for both the nick substrate and double-stranded DNA might be the same.
- Biological Information Research Center and the Gene Discovery Research Center, National Institute of Advanced Industrial Science and Technology, Higashi 1-1-1, Tsukuba, Ibaraki 305-566, Japan.
Organizational Affiliation: