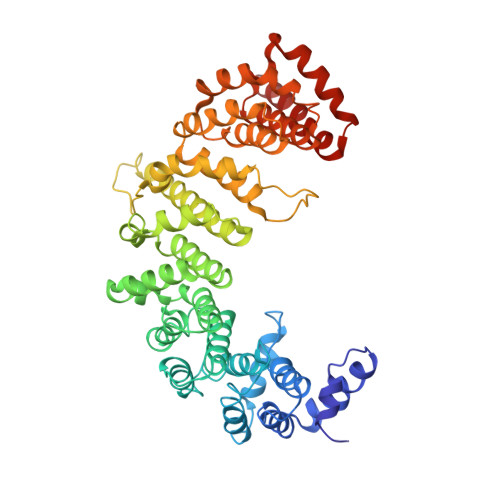
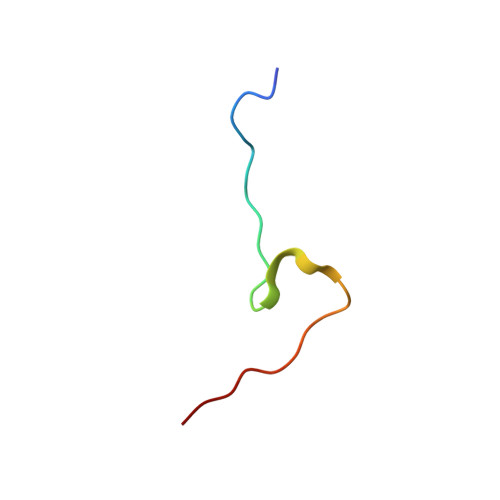
Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta
Cingolani, G., Bednenko, J., Gillespie, M.T., Gerace, L.(2002) Mol Cell 10: 1345-1353
- PubMed: 12504010
- DOI: https://doi.org/10.1016/s1097-2765(02)00727-x
- Primary Citation of Related Structures:
1M5N - PubMed Abstract:
Nuclear import of proteins containing a classical nuclear localization signal (NLS) involves NLS recognition by importin alpha, which associates with importin beta via the IBB domain. Other proteins, including parathyroid hormone-related protein (PTHrP), are imported into the nucleus by direct interaction with importin beta. We solved the crystal structure of a fragment of importin beta-1 (1-485) bound to the nonclassical NLS of PTHrP. The structure reveals a second extended cargo binding site on importin beta distinct from the IBB domain binding site. Using a permeabilized cell import assay we demonstrate that importin beta (1-485) can import PTHrP-coupled cargo in a Ran-dependent manner. We propose that this region contains a prototypical nuclear import receptor domain, which could have evolved into the modern importin beta superfamily.
- Departments of Cell and Molecular Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: