Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.
Jormakka, M., Tornroth, S., Byrne, B., Iwata, S.(2002) Science 295: 1863-1868
- PubMed: 11884747
- DOI: https://doi.org/10.1126/science.1068186
- Primary Citation of Related Structures:
1KQF, 1KQG - PubMed Abstract:
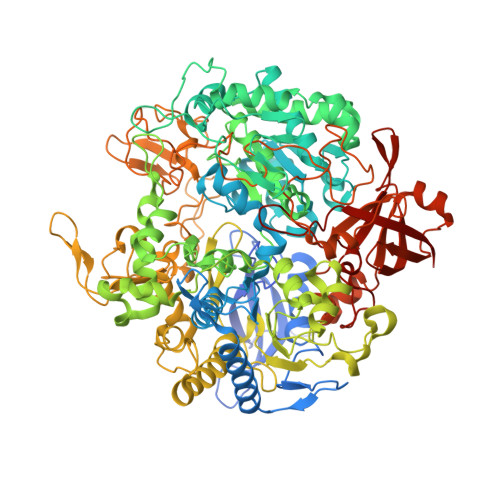
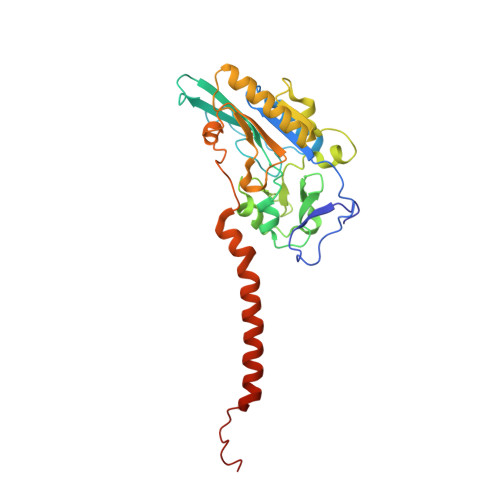
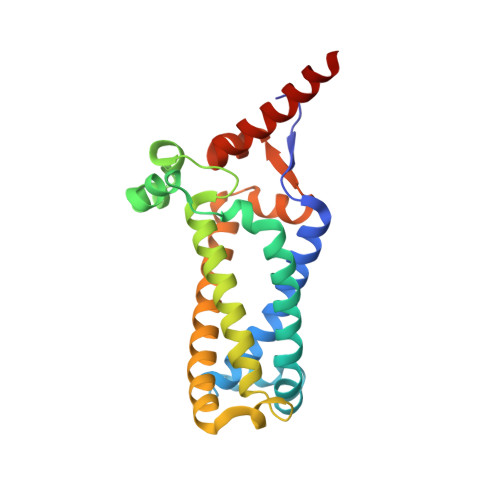
The structure of the membrane protein formate dehydrogenase-N (Fdn-N), a major component of Escherichia coli nitrate respiration, has been determined at 1.6 angstroms. The structure demonstrates 11 redox centers, including molybdopterin-guanine dinucleotides, five [4Fe-4S] clusters, two heme b groups, and a menaquinone analog. These redox centers are aligned in a single chain, which extends almost 90 angstroms through the enzyme. The menaquinone reduction site associated with a possible proton pathway was also characterized. This structure provides critical insights into the proton motive force generation by redox loop, a common mechanism among a wide range of respiratory enzymes.
- Division of Biomedical Sciences, Imperial College, London SW7 2AZ, UK.
Organizational Affiliation: