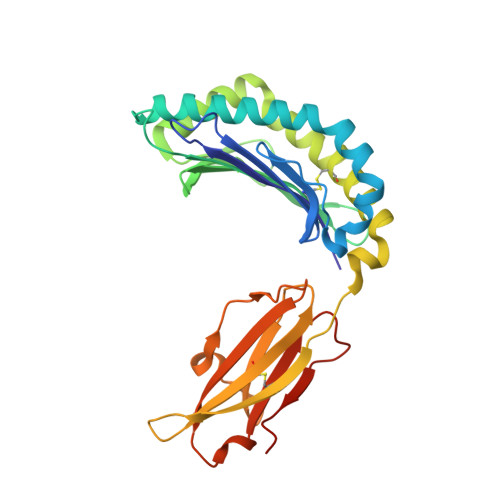
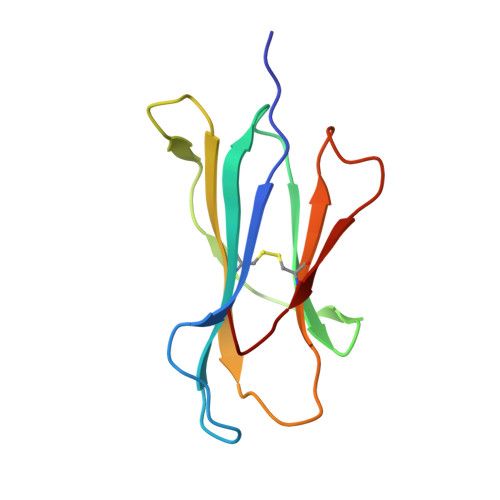
Crystal structures of two rat MHC class Ia (RT1-A) molecules that are associated differentially with peptide transporter alleles TAP-A and TAP-B.
Rudolph, M.G., Stevens, J., Speir, J.A., Trowsdale, J., Butcher, G.W., Joly, E., Wilson, I.A.(2002) J Mol Biology 324: 975-990
- PubMed: 12470953
- DOI: https://doi.org/10.1016/s0022-2836(02)01095-1
- Primary Citation of Related Structures:
1KJM, 1KJV - PubMed Abstract:
Antigenic peptides are loaded onto class I MHC molecules in the endoplasmic reticulum (ER) by a complex consisting of the MHC class I heavy chain, beta(2)-microglobulin, calreticulin, tapasin, Erp57 (ER60) and the transporter associated with antigen processing (TAP). While most mammalian species transport these peptides into the ER via a single allele of TAP, rats have evolved different TAPs, TAP-A and TAP-B, that are present in different inbred strains. Each TAP delivers a different spectrum of peptides and is associated genetically with distinct subsets of MHC class Ia alleles, but the molecular basis for the conservation (or co-evolution) of the two transporter alleles is unknown. We have determined the crystal structures of a representative of each MHC subset, viz RT1-A(a) and RT1-A1(c), in association with high-affinity nonamer peptides. The structures reveal how the chemical properties of the two different rat MHC F-pockets match those of the corresponding C termini of the peptides, corroborating biochemical data on the rates of peptide-MHC complex assembly. An unusual sequence in RT1-A1(c) leads to a major deviation from the highly conserved beta(3)/alpha(1) loop (residues 40-59) conformation in mouse and human MHC class I structures. This loop change contributes to profound changes in the shape of the A-pocket in the peptide-binding groove and may explain the function of RT1-A1(c) as an inhibitory natural killer cell ligand.
- Department of Molecular Biology and Skaggs Institute for Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: