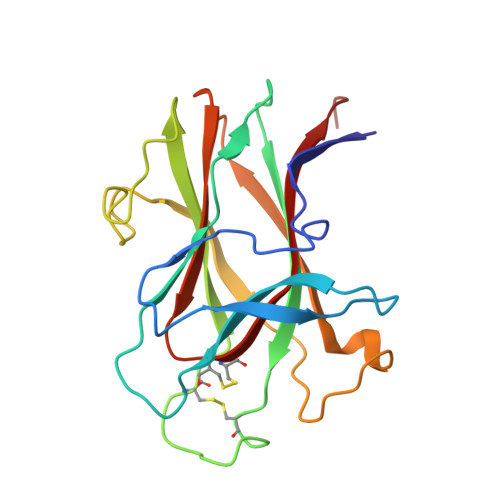
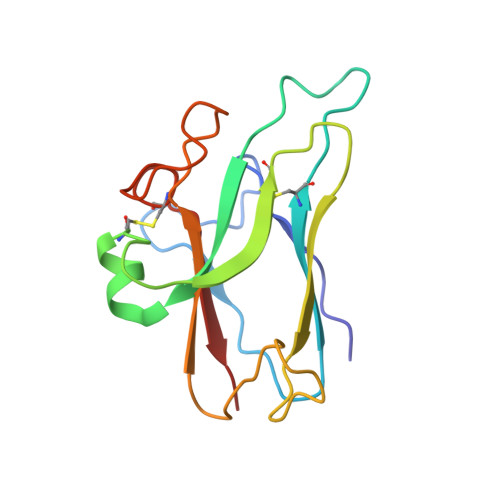
Crystal structure of an Eph receptor-ephrin complex.
Himanen, J.P., Rajashankar, K.R., Lackmann, M., Cowan, C.A., Henkemeyer, M., Nikolov, D.B.(2001) Nature 414: 933-938
- PubMed: 11780069
- DOI: https://doi.org/10.1038/414933a
- Primary Citation of Related Structures:
1KGY - PubMed Abstract:
The Eph family of receptor tyrosine kinases and their membrane-anchored ephrin ligands are important in regulating cell-cell interactions as they initiate a unique bidirectional signal transduction cascade whereby information is communicated into both the Eph-expressing and the ephrin-expressing cells. Initially identified as regulators of axon pathfinding and neuronal cell migration, Ephs and ephrins are now known to have roles in many other cell-cell interactions, including those of vascular endothelial cells and specialized epithelia. Here we report the crystal structure of the complex formed between EphB2 and ephrin-B2, determined at 2.7 A resolution. Each Eph receptor binds an ephrin ligand through an expansive dimerization interface dominated by the insertion of an extended ephrin loop into a channel at the surface of the receptor. Two Eph-Ephrin dimers then join to form a tetramer, in which each ligand interacts with two receptors and each receptor interacts with two ligands. The Eph and ephrin molecules are precisely positioned and orientated in these complexes, promoting higher-order clustering and the initiation of bidirectional signalling.
- Cellular Biochemistry and Biophysics Program, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, New York 10021, USA.
Organizational Affiliation: