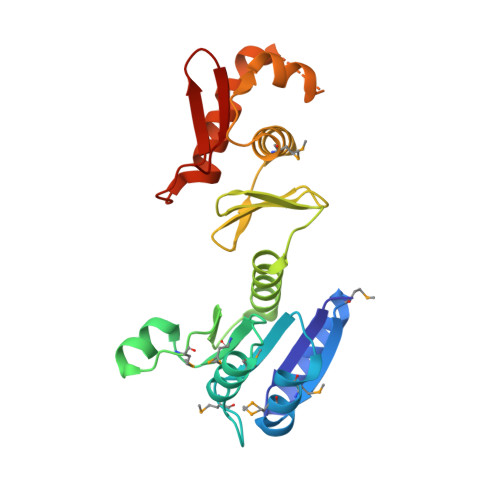
Evidence of intradomain and interdomain flexibility in an OmpR/PhoB homolog from Thermotoga maritima.
Buckler, D.R., Zhou, Y., Stock, A.M.(2002) Structure 10: 153-164
- PubMed: 11839301
- DOI: https://doi.org/10.1016/s0969-2126(01)00706-7
- Primary Citation of Related Structures:
1KGS - PubMed Abstract:
Two-component systems, the predominant signal transduction strategy used by prokaryotes, involve phosphorelay from a sensor histidine kinase (HK) to an intracellular response regulator protein (RR) that typically acts as a transcription regulator. RRs are modular proteins, usually composed of a conserved regulatory domain, which functions as a phosphorylation-activated switch, and an attached DNA binding effector domain. The crystal structure of a Thermotoga maritima transcription factor, DrrD, has been determined at 1.5 A resolution, providing the first structural information for a full-length member of the OmpR/PhoB subfamily of RRs. A small interdomain interface occurs between alpha 5 of the regulatory domain and an antiparallel sheet of the effector domain. The lack of an extensive interface in the unphosphorylated protein distinguishes DrrD from other structurally characterized multidomain RRs and suggests a different mode of interdomain regulation.
- Center for Advanced Biotechnology and Medicine, 679 Hoes Lane, Piscataway, NJ 08854, USA.
Organizational Affiliation: