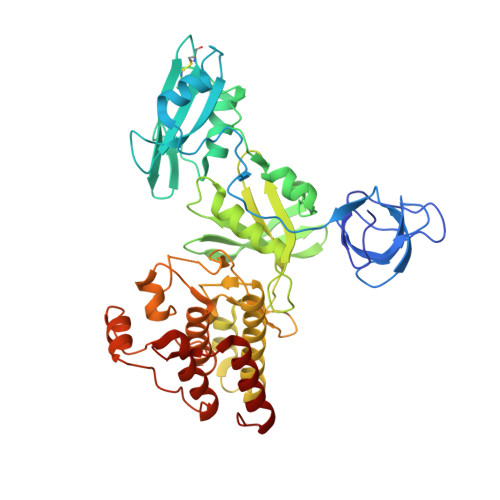
Structure of the carboxyl-terminal Src kinase, Csk.
Ogawa, A., Takayama, Y., Sakai, H., Chong, K.T., Takeuchi, S., Nakagawa, A., Nada, S., Okada, M., Tsukihara, T.(2002) J Biological Chem 277: 14351-14354
- PubMed: 11884384
- DOI: https://doi.org/10.1074/jbc.C200086200
- Primary Citation of Related Structures:
1K9A - PubMed Abstract:
The carboxyl-terminal Src kinase (Csk) is an indispensable negative regulator for the Src family tyrosine kinases (SFKs) that play pivotal roles in various cell signalings. To understand the molecular basis of the Csk-mediated regulation of SFKs, we elucidated the crystal structure of full-length Csk. The Csk crystal consists of six molecules classified as active or inactive states according to the coordinations of catalytic residues. Csk assembles the SH2 and SH3 domains differently from inactive SFKs, and their binding pockets are oriented outward enabling the intermolecular interaction. In active molecules, the SH2-kinase and SH2-SH3 linkers are tightly stuck to the N-lobe of the kinase domain to stabilize the active conformation, and there is a direct linkage between the SH2 and the kinase domains. In inactive molecules, the SH2 domains are rotated destroying the linkage to the kinase domain. Cross-correlation matrices for the active molecules reveal that the SH2 domain and the N-lobe of the kinase domain move as a unit. These observations suggest that Csk can be regulated through coupling of the SH2 and kinase domains and that Csk provides a novel built-in activation mechanism for cytoplasmic tyrosine kinases.
- Institute for Protein Research, Osaka University, 3-2 Yamadaoka and Research Institute for Microbial Diseases, Osaka University, 3-1 Yamadaoka, Suita, Osaka 565-0871, Japan.
Organizational Affiliation: