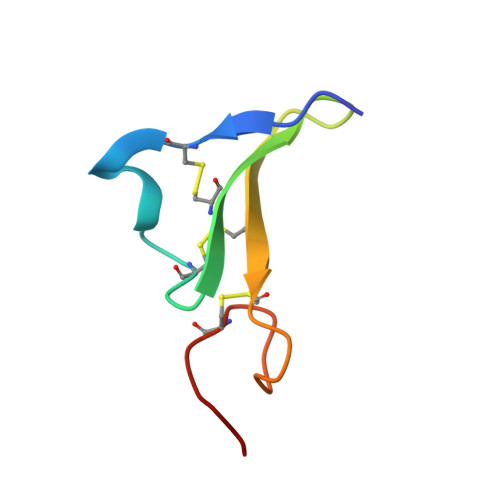
Solution structure of epiregulin and the effect of its C-terminal domain for receptor binding affinity
Sato, K., Nakamura, T., Mizuguchi, M., Miura, K., Tada, M., Aizawa, T., Gomi, T., Miyamoto, K., Kawano, K.(2003) FEBS Lett 553: 232-238
- PubMed: 14572630
- DOI: https://doi.org/10.1016/s0014-5793(03)01005-6
- Primary Citation of Related Structures:
1K36, 1K37 - PubMed Abstract:
Epiregulin (EPR), a novel member of epidermal growth factor (EGF) family, is a ligand for ErbB-1 and ErbB-4 receptors. The binding affinity of EPR for the receptors is lower than those of other EGF-family ligands. The solution structure of EPR was determined using two-dimensional nuclear magnetic resonance spectroscopy. The secondary structure in the C-terminal domain of EPR is different from other EGF-family ligands because of the lack of hydrogen bonds. The structural difference in the C-terminal domain may provide an explanation for the reduced binding affinity of EPR to the ErbB receptors.
- Faculty of Pharmaceutical Sciences, Toyama Medical and Pharmaceutical University, 2630 Sugitani, 930-0194 Toyama, Japan.
Organizational Affiliation: