The role of beta chains in the control of the hemoglobin oxygen binding function: chimeric human/mouse proteins, structure, and function.
Kidd, R.D., Russell, J.E., Watmough, N.J., Baker, E.N., Brittain, T.(2001) Biochemistry 40: 15669-15675
- PubMed: 11747442
- DOI: https://doi.org/10.1021/bi011329f
- Primary Citation of Related Structures:
1JEB - PubMed Abstract:
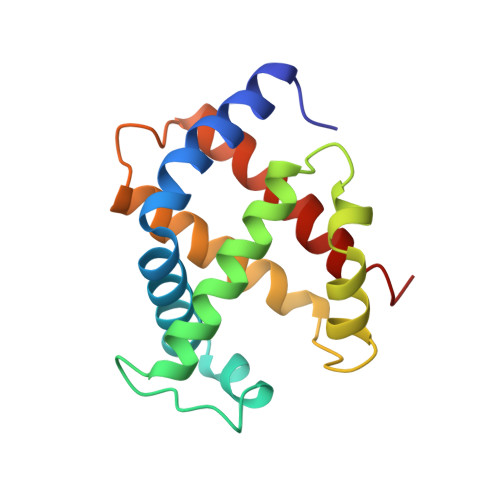
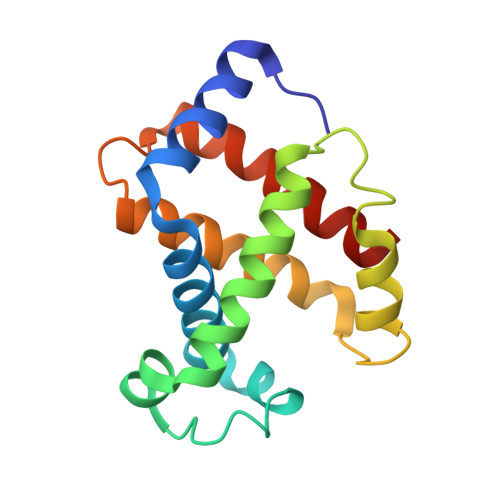
By using transgenic methodologies, we have produced a number of mouse/human chimeric hemoglobins containing adult mouse and human embryonic globin chains. A detailed analysis of the oxygen binding properties of these proteins identifies the dominant role played by the specific beta-type globin chains in the control of the oxygen binding characteristics. Further analysis traces the origins of these effects to alterations in the properties of the T states of these proteins. The human zeta/mouse beta chimeric protein has been crystallized, and its structure has been determined by X-ray diffraction to a resolution of 2.1 A with R (R(free)) values of 21.6% (24.9%). Close examination of the structure indicates that the subunit interfaces contain contacts which, although different from those present in either the parent human or the parent mouse proteins, retain the overall stabilizing interactions seen in other R state hemoglobins.
- School of Biological Sciences, Private Bag 92019, University of Auckland, Auckland, New Zealand.
Organizational Affiliation: