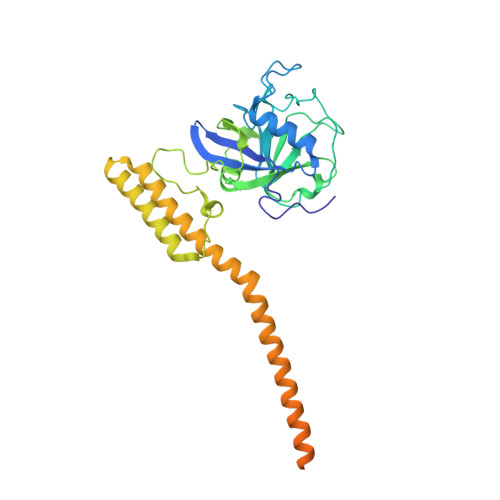
Two structures of cyclophilin 40: folding and fidelity in the TPR domains.
Taylor, P., Dornan, J., Carrello, A., Minchin, R.F., Ratajczak, T., Walkinshaw, M.D.(2001) Structure 9: 431-438
- PubMed: 11377203
- DOI: https://doi.org/10.1016/s0969-2126(01)00603-7
- Primary Citation of Related Structures:
1IHG, 1IIP - PubMed Abstract:
The "large immunophilin" family consists of domains of cyclophilin or FK506 binding protein linked to a tetratricopeptide (TPR) domain. They are intimately associated with steroid receptor complexes and bind to the C-terminal domain of Hsp90 via the TPR domain. The competitive binding of specific large immunophilins and other TPR-Hsp90 proteins provides a regulatory mechanism for Hsp90 chaperone activity. We have solved the X-ray structures of monoclinic and tetragonal forms of Cyp40. In the monoclinic form, the TPR domain consists of seven helices of variable length incorporating three TPR motifs, which provide a convincing binding surface for the Hsp90 C-terminal MEEVD sequence. The C-terminal residues of Cyp40 protrude out beyond the body of the TPR domain to form a charged helix-the putative calmodulin binding site. However, in the tetragonal form, two of the TPR helices have straightened out to form one extended helix, providing a dramatically different conformation of the molecule. The X-ray structures are consistent with the role of Cyclophilin 40 as a multifunctional signaling protein involved in a variety of protein-protein interactions. The intermolecular helix-helix interactions in the tetragonal form mimic the intramolecular interactions found in the fully folded monoclinic form. These conserved intra- and intermolecular TPR-TPR interactions are illustrative of a high-fidelity recognition mechanism. The two structures also open up the possibility that partially folded forms of TPR may be important in domain swapping and protein recognition.
- Structural Biochemistry Group, Institute of Cell and Molecular Biology, The University of Edinburgh, Michael Swann Building, King's Buildings, Mayfield Road, EH9 3JR, Edinburgh, United Kingdom.
Organizational Affiliation: