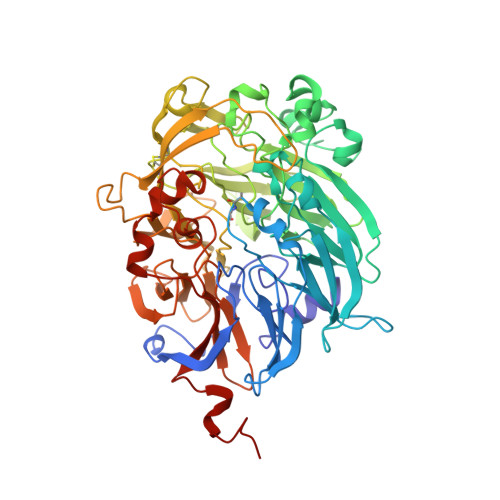
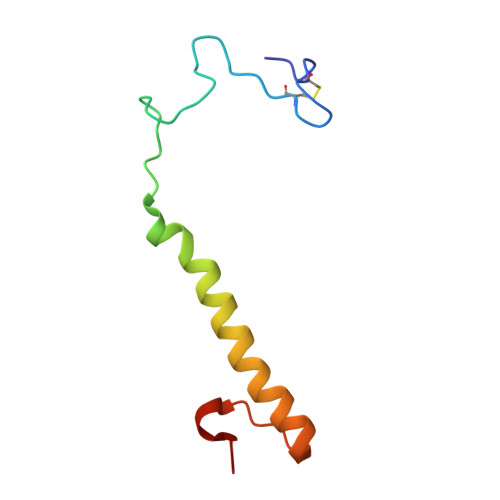
The Refined Structure of the Quinoprotein Methanol Dehydrogenase from Methylobacterium Extorquens at 1.94 A.
Ghosh, M., Anthony, C., Harlos, K., Goodwin, M.G., Blake, C.(1995) Structure 3: 177
- PubMed: 7735834
- DOI: https://doi.org/10.1016/s0969-2126(01)00148-4
- Primary Citation of Related Structures:
1H4I - PubMed Abstract:
Methanol dehydrogenase (MDH) is a bacterial periplasmic quinoprotein; it has pyrrolo-quinoline quinone (PQQ) as its prosthetic group, requires Ca2+ for activity and uses cytochrome cL as its electron acceptor. Low-resolution structures of MDH have already been determined. The structure of the alpha 2 beta 2 tetramer of MDH from Methylobacterium extorquens has now been determined at 1.94 A with an R-factor of 19.85%. The alpha-subunit of MDH has an eight-fold radial symmetry, with its eight beta-sheets stabilized by a novel tryptophan docking motif. The PQQ in the active site is held in place by a coplanar tryptophan and by a novel disulphide ring formed between adjacent cysteines which are bonded by an unusual non-planar trans peptide bond. One of the carbonyl oxygens of PQQ is bonded to the Ca2+, probably facilitating attack on the substrate, and the other carbonyl oxygen is out of the plane of the ring, confirming the presence of the predicted free-radical semiquinone form of the prosthetic group.
- Laboratory of Molecular Biophysics, University of Oxford, UK.
Organizational Affiliation: