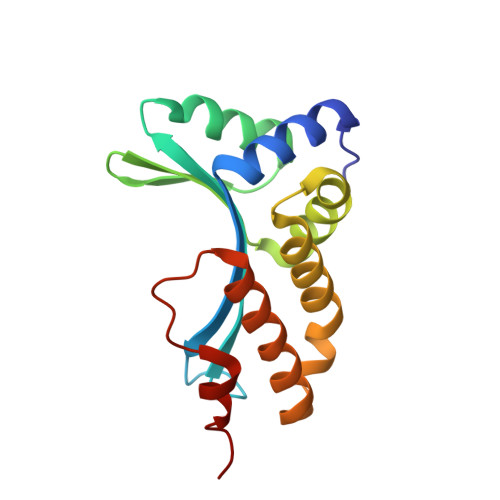
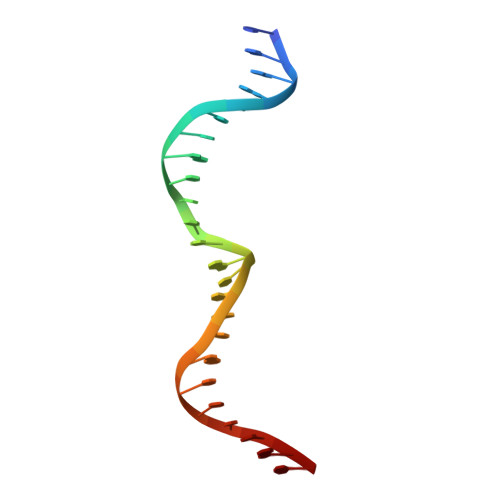
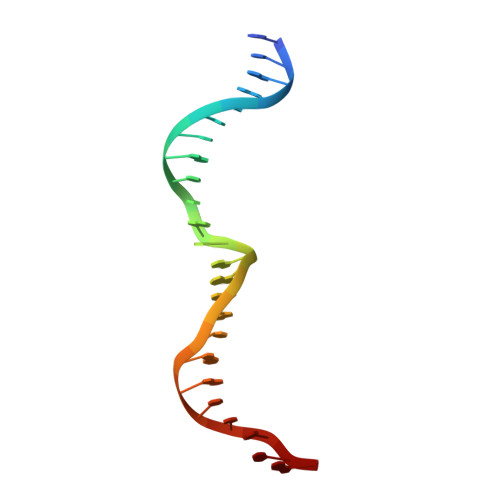
The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites.
Chevalier, B.S., Monnat Jr., R.J., Stoddard, B.L.(2001) Nat Struct Biol 8: 312-316
- PubMed: 11276249
- DOI: https://doi.org/10.1038/86181
- Primary Citation of Related Structures:
1G9Y, 1G9Z - PubMed Abstract:
Homing endonucleases, like restriction enzymes, cleave double-stranded DNA at specific target sites. The cleavage mechanism(s) utilized by LAGLIDADG endonucleases have been difficult to elucidate; their active sites are divergent, and only one low resolution cocrystal structure has been determined. Here we report two high resolution structures of the dimeric I-CreI homing endonuclease bound to DNA: a substrate complex with calcium and a product complex with magnesium. The bound metals in both complexes are verified by manganese anomalous difference maps. The active sites are positioned close together to facilitate cleavage across the DNA minor groove; each contains one metal ion bound between a conserved aspartate (Asp 20) and a single scissile phosphate. A third metal ion bridges the two active sites. This divalent cation is bound between aspartate residues from the active site of each subunit and is in simultaneous contact with the scissile phosphates of both DNA strands. A metal-bound water molecule acts as the nucleophile and is part of an extensive network of ordered water molecules that are positioned by enzyme side chains. These structures illustrate a unique variant of a two-metal endonuclease mechanism is employed by the highly divergent LAGLIDADG enzyme family.
- Fred Hutchinson Cancer Research Center, University of Washington, 1100 Fairview Ave. N. A3-023, Seattle, Washington 98109, USA.
Organizational Affiliation: