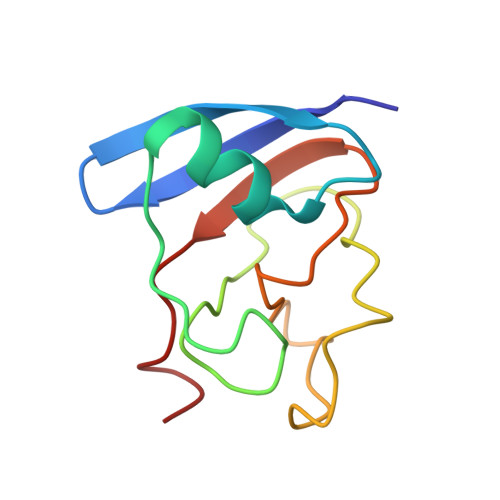
Structure of the [2Fe-2S] ferredoxin I from the blue-green alga Aphanothece sacrum at 2.2 A resolution.
Tsukihara, T., Fukuyama, K., Mizushima, M., Harioka, T., Kusunoki, M., Katsube, Y., Hase, T., Matsubara, H.(1990) J Mol Biology 216: 399-410
- PubMed: 2123937
- DOI: https://doi.org/10.1016/S0022-2836(05)80330-4
- Primary Citation of Related Structures:
1FXI - PubMed Abstract:
Crystals of a [2Fe-2S] ferredoxin (Fd) I with a relative molecular mass of 10,480 were obtained from the blue-green alga Aphanothece sacrum. Each asymmetric unit of the crystal contains four molecules. An electron density map calculated by the single isomorphous replacement method with the anomalous dispersion at 2.5 A resolution was refined by averaging the four molecules in the asymmetric unit. Positional and isotropic thermal parameters for the non-hydrogen atoms of the four molecules and 158 water molecules were refined to an R-factor (R = sigma[Fo-Fc[/sigma Fo) of 0.23 by the restrained least-squares method. The estimated root-mean-square (r.m.s.) error for the atomic positions is 0.3 A. The r.m.s. deviations of equivalent C alpha atoms of the asymmetric-unit molecules superposed by the least-squares method average 0.35 A. The Fd molecule has a structure like the beta-barrel in the molecule of the [2Fe-2S] Fd from Spirulina platensis. A [2Fe-2S] cluster is bonded covalently to the protein molecule by four Fe-S, in which three of the Fe-S bonds are in a loop segment from position 38 to 47. The hydrophobic core inside the beta-barrel is formed by seven conservative residues: Val15, Val18, Ile24, Leu51, Ile74, Ala79 and Ile87. The molecular surface around Tyr23, Tyr80 and the active center may interact with ferredoxin-NADP+ reductase. One of the two iron atoms of the [2Fe-2S] cluster should be more easily reduced than the other because of differences in the hydrogen-bonding scheme and the hydrophobicity around the atoms.
- Faculty of Engineering, Tottori University, Japan.
Organizational Affiliation: