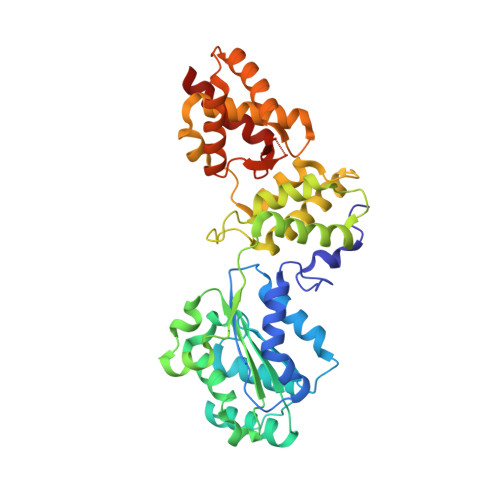
Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control.
Liu, J., Smith, C.L., DeRyckere, D., DeAngelis, K., Martin, G.S., Berger, J.M.(2000) Mol Cell 6: 637-648
- PubMed: 11030343
- DOI: https://doi.org/10.1016/s1097-2765(00)00062-9
- Primary Citation of Related Structures:
1FNN - PubMed Abstract:
Cdc6/Cdc18 is a conserved and essential component of prereplication complexes. The 2.0 A crystal structure of an archaeal Cdc6 ortholog, in conjunction with a mutational analysis of the homologous Cdc18 protein from Schizosaccharomyces pombe, reveals novel aspects of Cdc6/Cdc18 function. Two domains of Cdc6 form an AAA+-type nucleotide binding fold that is observed bound to Mg.ADP. A third domain adopts a winged-helix fold similar to known DNA binding modules. Sequence comparisons show that the winged-helix domain is conserved in Orc1, and mutagenesis data demonstrate that this region of Cdc6/Cdc18 is required for function in vivo. Additional mutational analyses suggest that nucleotide binding and/or hydrolysis by Cdc6/Cdc18 is required not only for progression through S phase, but also for maintenance of checkpoint control during S phase.
- Department of Molecular and Cell Biology, University of California, Berkeley 94720, USA.
Organizational Affiliation: