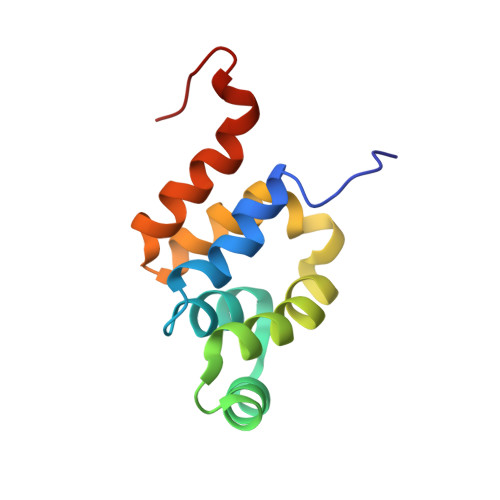
The Three-Dimensional Solution Structure and Dynamic Properties of the Human Fadd Death Domain
Berglund, H., Olerenshaw, D., Sankar, A., Federwisch, M., Mcdonald, N.Q., Driscoll, P.C.(2000) J Mol Biology 302: 171
- PubMed: 10964568
- DOI: https://doi.org/10.1006/jmbi.2000.4011
- Primary Citation of Related Structures:
1E3Y, 1E41 - PubMed Abstract:
FADD (also known as MORT-1) is an essential adapter protein that couples the transmembrane receptors Fas (CD95) and tumor necrosis factor receptor-1 (TNF-R1) to intracellular cysteine proteases known as caspases, which propagate and execute the programmed cell death-inducing signal triggered by Fas ligand (FasL, CD95L) and TNF. FADD contains 208 amino acid residues, and comprises two functionally and structurally distinct domains: an N-terminal death effector domain (DED) that promotes activation of the downstream proteolytic cascade through binding of the DED domains of procaspase-8; and a C-terminal death domain (DD). FADD-DD provides the site of FADD recruitment to death receptor complexes at the plasma membrane by, for example, interaction with the Fas receptor cytoplasmic death domain (Fas-DD), or binding of the TNF-R1 adapter molecule TRADD. We have determined the three-dimensional solution structure and characterised the internal polypeptide dynamics of human FADD-DD using heteronuclear NMR spectroscopy of (15)N and (13)C,(15)N-labelled samples. The structure comprises six alpha-helices joined by short loops and displays overall similarity to the death domain of the Fas receptor. The analysis of the dynamic properties reveals no evidence of contiguous stretches of polypeptide chain with increased internal motion, except at the extreme chain termini. A pattern of increased rates of amide proton solvent exchange in the alpha3 helix correlates with a higher degree of solvent exposure for this secondary structure element. The properties of the FADD-DD structure are discussed with respect to previously reported mutagenesis data and emerging models for FasL-induced FADD recruitment to Fas and caspase-8 activation.
- Department of Biochemistry and Molecular Biology, University College London, Gower Street, London, WC1E 6BT, UK.
Organizational Affiliation: