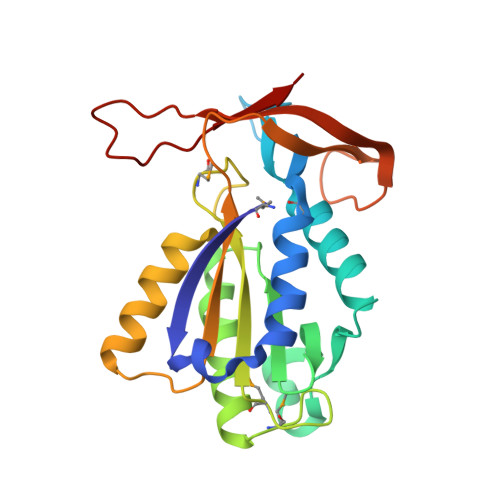
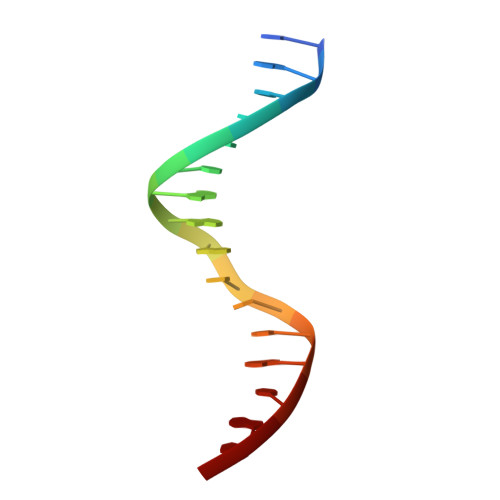
Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution.
Lukacs, C.M., Kucera, R., Schildkraut, I., Aggarwal, A.K.(2000) Nat Struct Biol 7: 134-140
- PubMed: 10655616
- DOI: https://doi.org/10.1038/72405
- Primary Citation of Related Structures:
1D2I, 1DFM - PubMed Abstract:
Restriction endonucleases are remarkably resilient to alterations in their DNA binding specificity. To understand the basis of this immutability, we have determined the crystal structure of endonuclease BglII bound to its recognition sequence (AGATCT), at 1. 5 A resolution. We compare the structure of BglII to endonuclease BamHI, which recognizes a closely related DNA site (GGATCC). We show that both enzymes share a similar alpha/beta core, but in BglII, the core is augmented by a beta-sandwich domain that encircles the DNA to provide extra specificity. Remarkably, the DNA is contorted differently in the two structures, leading to different protein-DNA contacts for even the common base pairs. Furthermore, the BglII active site contains a glutamine in place of the glutamate at the general base position in BamHI, and only a single metal is found coordinated to the putative nucleophilic water and the phosphate oxygens. This surprising diversity in structures shows that different strategies can be successful in achieving site-specific recognition and catalysis in restriction endonucleases.
- Structural Biology Program, Department of Physiology and Biophysics, Mt. Sinai School of Medicine, 1425 Madison Avenue, New York, New York 10029, USA.
Organizational Affiliation: