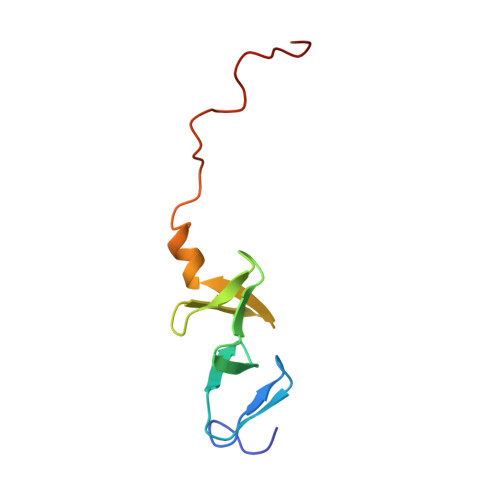
Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP.
Perez-Alvarado, G.C., Miles, C., Michelsen, J.W., Louis, H.A., Winge, D.R., Beckerle, M.C., Summers, M.F.(1994) Nat Struct Biol 1: 388-398
- PubMed: 7664053
- DOI: https://doi.org/10.1038/nsb0694-388
- Primary Citation of Related Structures:
1CTL - PubMed Abstract:
The three dimensional solution structure of the carboxy terminal LIM domain of the avian Cysteine Rich Protein (CRP) has been determined by nuclear magnetic resonance spectroscopy. The domain contains two zinc atoms bound independently in CCHC (C = Cys, H = His) and CCCC modules. Both modules contain two orthogonally-arranged antiparallel beta-sheets, and the CCCC module contains an alpha-helix at its C terminus. The modules pack due to hydrophobic interactions forming a novel global fold. The structure of the C-terminal CCCC module is essentially identical to that observed for the DNA-interactive CCCC modules of the GATA-1 and steroid hormone receptor DNA binding domains, raising the possibility that the LIM motif may have a DNA binding function.
- Howard Hughes Medical Institute, University of Maryland Baltimore County 21228, USA.
Organizational Affiliation: