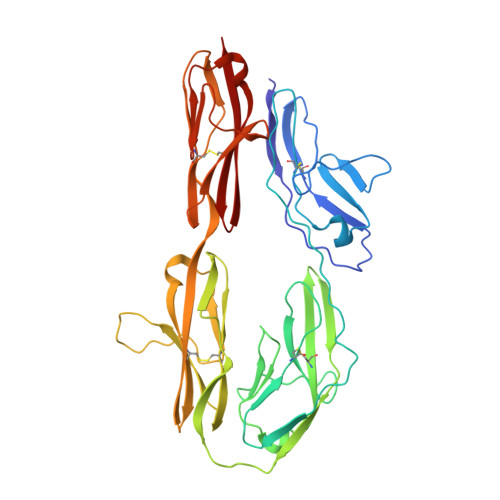
The crystal structure of the ligand binding module of axonin-1/TAG-1 suggests a zipper mechanism for neural cell adhesion.
Freigang, J., Proba, K., Leder, L., Diederichs, K., Sonderegger, P., Welte, W.(2000) Cell 101: 425-433
- PubMed: 10830169
- DOI: https://doi.org/10.1016/s0092-8674(00)80852-1
- Primary Citation of Related Structures:
1CS6 - PubMed Abstract:
We have determined the crystal structure of the ligand binding fragment of the neural cell adhesion molecule axonin-1/TAG-1 comprising the first four immunoglobulin (Ig) domains. The overall structure of axonin-1(Ig1-4) is U-shaped due to contacts between domains 1 and 4 and domains 2 and 3. In the crystals, these molecules are aligned in a string with adjacent molecules oriented in an anti-parallel fashion and their C termini perpendicular to the string. This arrangement suggests that cell adhesion by homophilic axonin-1 interaction occurs by the formation of a linear zipper-like array in which the axonin-1 molecules are alternately provided by the two apposed membranes. In accordance with this model, mutations in a loop critical for the formation of the zipper resulted in the loss of the homophilic binding capacity of axonin-1.
- Faculty of Biology, University of Konstanz, Germany.
Organizational Affiliation: