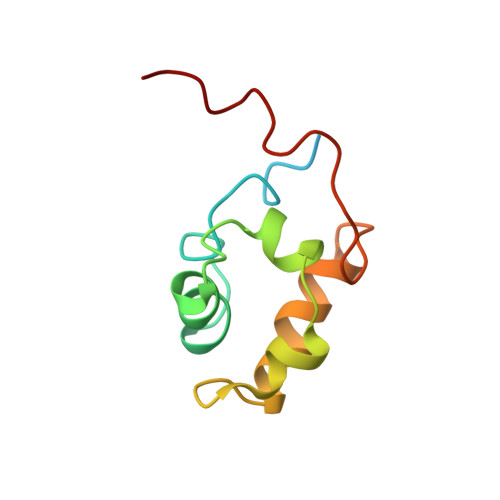
Solution structure of the activator contact domain of the RNA polymerase alpha subunit.
Jeon, Y.H., Negishi, T., Shirakawa, M., Yamazaki, T., Fujita, N., Ishihama, A., Kyogoku, Y.(1995) Science 270: 1495-1497
- PubMed: 7491496
- DOI: https://doi.org/10.1126/science.270.5241.1495
- Primary Citation of Related Structures:
1COO - PubMed Abstract:
The structure of the carboxyl-terminal domain of the Escherichia coli RNA polymerase alpha subunit (alpha CTD), which is regarded as the contact site for transcription activator proteins and for the promoter UP element, was determined by nuclear magnetic resonance spectroscopy. Its compact structure of four helices and two long arms enclosing its hydrophobic core shows a folding topology distinct from those of other DNA-binding proteins. The UP element binding site was found on the surface comprising helix 1, the amino-terminal end of helix 4, and the preceding loop. Mutation experiments indicated that the contact sites for transcription activator proteins are also on the same surface.
- Institute for Protein Research, Osaka University, Japan.
Organizational Affiliation: