Structures of escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity.
Briozzo, P., Golinelli-Pimpaneau, B., Gilles, A.M., Gaucher, J.F., Burlacu-Miron, S., Sakamoto, H., Janin, J., Barzu, O.(1998) Structure 6: 1517-1527
- PubMed: 9862805
- DOI: https://doi.org/10.1016/s0969-2126(98)00150-6
- Primary Citation of Related Structures:
1CKE, 2CMK - PubMed Abstract:
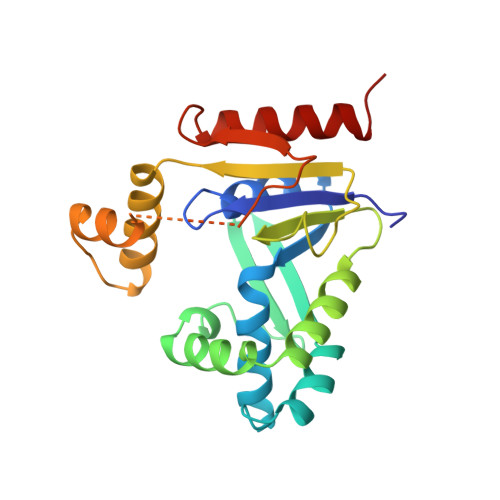
. Nucleoside monophosphate kinases (NMP kinases) catalyze the reversible transfer of a phosphoryl group from a nucleoside triphosphate to a nucleoside monophosphate. Among them, cytidine monophosphate kinase from Escherichia coli has a striking particularity: it is specific for CMP, whereas in eukaryotes a unique UMP/CMP kinase phosphorylates both CMP and UMP with similar efficiency. . The crystal structure of the CMP kinase apoenzyme from E. coli was solved by single isomorphous replacement and refined at 1.75 A resolution. The structure of the enzyme in complex with CDP was determined at 2.0 A resolution. Like other NMP kinases, the protein contains a central parallel beta sheet, the strands of which are connected by alpha helices. The enzyme differs from other NMP kinases in the presence of a 40-residue insert situated in the NMP-binding (NMPbind) domain. This insert contains two domains: one comprising a three-stranded antiparallel beta sheet, the other comprising two alpha helices. . Two features of the CMP kinase from E. coli have no equivalent in other NMP kinases of known structure. Firstly, the large NMPbind insert undergoes a CDP-induced rearrangement: its beta-sheet domain moves away from the substrate, whereas its helical domain comes closer to it in a motion likely to improve the protection of the active site. Secondly, residues involved in CDP recognition are conserved in CMP kinases and have no counterpart in other NMP kinases. The structures presented here are the first of a new family of NMP kinases specific for CMP.
- Laboratoire de Chimie Biologique Institut National Agronomique Paris-Grignon 78850 Thiverval-Grignon France. Pierre.Briozzo@lebs. cnrs-gif.fr
Organizational Affiliation: