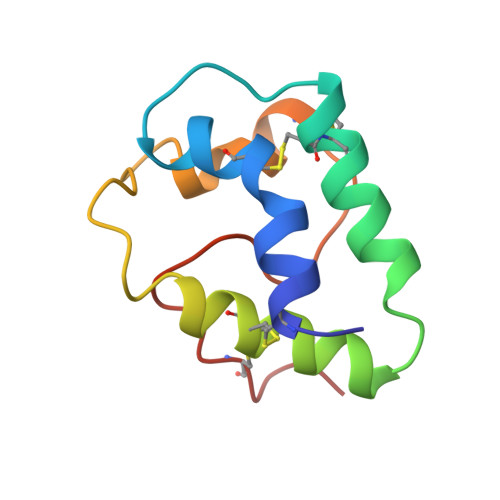
Solution structure of a lipid transfer protein extracted from rice seeds. Comparison with homologous proteins.
Poznanski, J., Sodano, P., Suh, S.W., Lee, J.Y., Ptak, M., Vovelle, F.(1999) Eur J Biochem 259: 692-708
- PubMed: 10092854
- DOI: https://doi.org/10.1046/j.1432-1327.1999.00093.x
- Primary Citation of Related Structures:
1BV2 - PubMed Abstract:
Nuclear magnetic resonance (NMR) spectroscopy was used to determine the three dimensional structure of rice nonspecific lipid transfer protein (ns-LTP), a 91 amino acid residue protein belonging to the broad family of plant ns-LTP. Sequence specific assignment was obtained for all but three HN backbone 1H resonances and for more than 95% of the 1H side-chain resonances using a combination of 1H 2D NOESY; TOCSY and COSY experiments at 293 K. The structure was calculated on the basis of four disulfide bridge restraints, 1259 distance constraints derived from 1H-1H Overhauser effects, 72 phi angle restraints and 32 hydrogen-bond restraints. The final solution structure involves four helices (H1: Cys3-Arg18, H2: Ala25-Ala37, H3: Thr41-Ala54 and H4: Ala66-Cys73) followed by a long C-terminal tail (T) with no observable regular structure. N-capping residues (Thr2, Ser24, Thr40), whose side-chain oxygen atoms are involved in hydrogen bonds with i + 3 amide proton additionally stabilize the N termini of the first three helices. The fourth helix involving Pro residues display a mixture of alpha and 3(10) conformation. The rms deviation of 14 final structures with respect to the average structure is 1.14 +/- 0.16 A for all heavy atoms (C, N, O and S) and 0.72 +/- 0.01 A for the backbone atoms. The global fold of rice ns-LTP is close to the previously published structures of wheat, barley and maize ns-LTPs exhibiting nearly identical pattern of the numerous sequence specific interactions. As reported previously for different four-helix topology proteins, hydrophobic, hydrogen bonding and electrostatic mechanisms of fold stabilization were found for the rice ns-LTP. The sequential alignment of 36 ns-LTP primary structures strongly suggests that there is a uniform pattern of specific long-range interactions (in terms of sequence), which stabilize the fold of all plant ns-LTPs.
- Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland.
Organizational Affiliation: