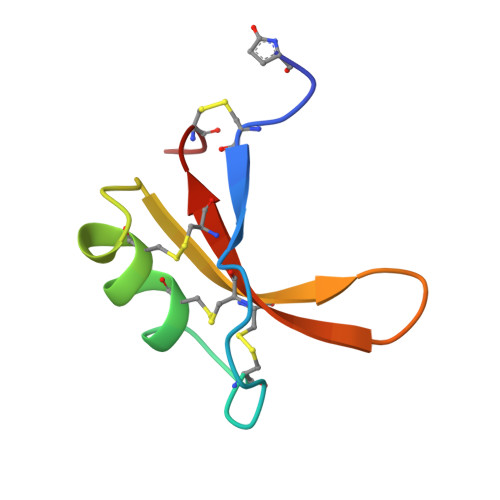
Solution structure of the thermostable sweet-tasting protein brazzein.
Caldwell, J.E., Abildgaard, F., Dzakula, Z., Ming, D., Hellekant, G., Markley, J.L.(1998) Nat Struct Biol 5: 427-431
- PubMed: 9628478
- DOI: https://doi.org/10.1038/nsb0698-427
- Primary Citation of Related Structures:
1BRZ, 2BRZ - PubMed Abstract:
The fruit of Pentadiplandra brazzeana Baillon contains a small, sweet-tasting protein named brazzein. The structure of brazzein in solution was determined by proton nuclear magnetic resonance spectroscopy at pH 5.2 and 22 degrees C. The brazzein fold, which contains one alpha-helix and three strands of antiparallel beta-sheet, does not resemble that of either of the other two sweet-tasting proteins with known structures, monellin and thaumatin. Instead, the structure of brazzein resembles those of plant gamma-thionins and defensins and arthropod toxins. Sequence comparisons predict that members of a newly-identified family of serine proteinase inhibitors share the brazzein fold.
- Graduate Program in Biophysics, University of Wisconsin-Madison 53706, USA.
Organizational Affiliation: