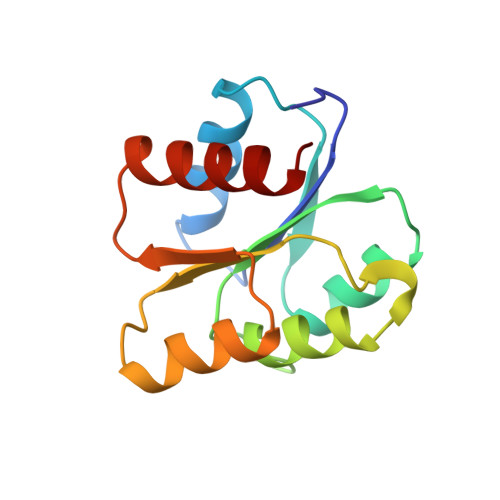
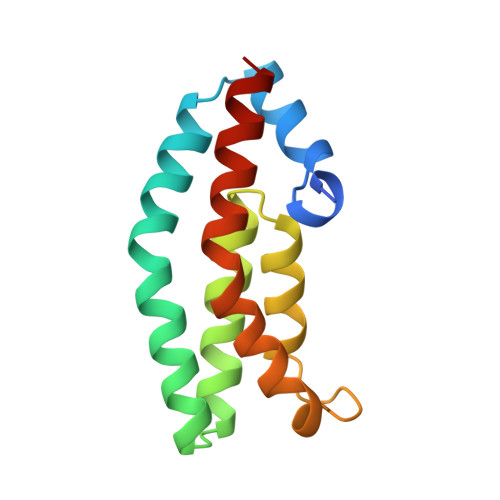
Structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor protein ArcB complexed with the chemotaxis response regulator CheY.
Kato, M., Shimizu, T., Mizuno, T., Hakoshima, T.(1999) Acta Crystallogr D Biol Crystallogr 55: 1257-1263
- PubMed: 10393292
- DOI: https://doi.org/10.1107/s0907444999005053
- Primary Citation of Related Structures:
1BDJ - PubMed Abstract:
The three-dimensional structure of the HPt domain of ArcB complexed with CheY has been determined using the molecular-replacement method. The structure was refined to a crystallographic R factor of 18.3% at 2.68 A resolution. The final model included 1899 protein atoms (117 residues from the HPt domain and 128 residues from CheY), one sulfate ion and 44 solvent molecules. In the crystal, CheY molecules stacked along the a axis of the cell with no interactions between neighbouring rows and the HPt domain bridged the CheY molecules. The phosphodonor residue His715 was fully exposed to the solvent region, even though the HPt domain was in contact with four molecules of CheY. CheY showed significant conformational change. This indicates that the HPt domain has a rigid structure when complexed with CheY.
- Department of Molecular Biology, Nara Institute of Science and Technology, 8916-5 Takayama Ikoma, Nara 630-0101, Japan.
Organizational Affiliation: