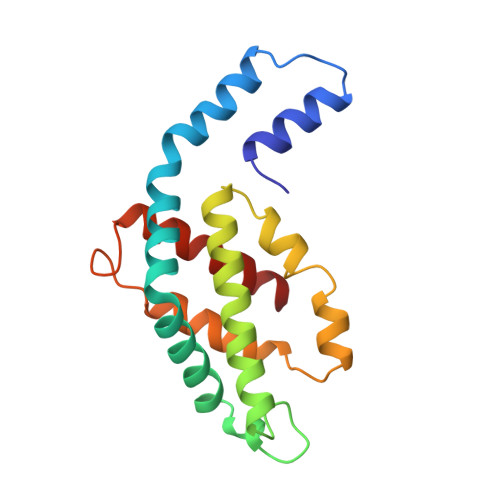
Crystal structure of a phycourobilin-containing phycoerythrin at 1.90-A resolution.
Ritter, S., Hiller, R.G., Wrench, P.M., Welte, W., Diederichs, K.(1999) J Struct Biol 126: 86-97
- PubMed: 10388620
- DOI: https://doi.org/10.1006/jsbi.1999.4106
- Primary Citation of Related Structures:
1B8D - PubMed Abstract:
The structure of R-phycoerythrin (R-PE) from the red alga Griffithsia monilis was solved at 1.90-A resolution by molecular replacement, using the atomic coordinates of cyanobacterial phycocyanin from Fremyella diplosiphon as a model. The crystallographic R factor for the final model is 17.5% (Rfree 22.7%) for reflections in the range 100-1.90 A. The model consists of an (alphabeta)2 dimer with an internal noncrystallographic dyad and a fragment of the gamma-polypeptide. The alpha-polypeptide (164 amino acid residues) has two covalently bound phycoerythrobilins at positions alpha82 and alpha139. The beta-polypeptide (177 residues) has two phycoerythrobilins bound to residues beta82 and beta158 and one phycourobilin covalently attached to rings A and D at residues beta50 and beta61, respectively. The electron density of the gamma-polypeptide is mostly averaged out by threefold crystallographic symmetry, but a dipeptide (Gly-Tyr) and one single Tyr could be modeled. These two tyrosine residues of the gamma-polypeptide are in close proximity to the phycoerythrobilins at position beta82 of two symmetry-related beta-polypeptides and are related by the same noncrystallographic dyad as the (alphabeta)2 dimer. Possible energy transfer pathways are discussed briefly.
- Institut für Biophysik und Strahlenbiologie, Universität Freiburg, Albertstrasse 23, Freiburg, D-79104, Germany.
Organizational Affiliation: