Full Text |
QUERY: Structure Author = "Dimattia, M.A." | MyPDB Login | Search API |
| Search Summary | This query matches 17 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry TypeSCOP Classification | 1 to 17 of 17 Structures Page 1 of 1 Sort by
Implications of the HIV-1 Rev dimer structure at 3.2A resolution for multimeric binding to the Rev response elementDiMattia, M.A., Watts, N.R., Stahl, S.J., Rader, C., Wingfield, P.T., Stuart, D.I., Steven, A.C., Grimes, J.M. (2010) Proc Natl Acad Sci U S A 107: 5810
Structural Characterization of Adeno-Associated Virus Serotype 9DiMattia, M.A., Nam, H.-J., Van Vliet, K., Mitchell, M., McCall, A., Bennett, A., Gurda, B., McKenna, R., Potter, M., Sakai, Y., Byrne, B.J., Muzyczka, N., Aslanidi, G., Zolotukhin, S., Olson, N., Sinkovitis, R., Baker, T.S., Agbandje-McKenna, M. (2012) J Virol 86: 6947-6958
Crystal Structure of an anti-HBV e-antigen monoclonal Fab fragment (e6), unboundDimattia, M.A., Watts, N.R., Stahl, S.J., Grimes, J.M., Steven, A.C., Stuart, D.I., Wingfield, P.T. (2013) Structure 21: 133-142
Crystal Structure of Hepatitis B Virus e-antigenDimattia, M.A., Watts, N.R., Stahl, S.J., Grimes, J.M., Steven, A.C., Stuart, D.I., Wingfield, P.T. (2013) Structure 21: 133-142
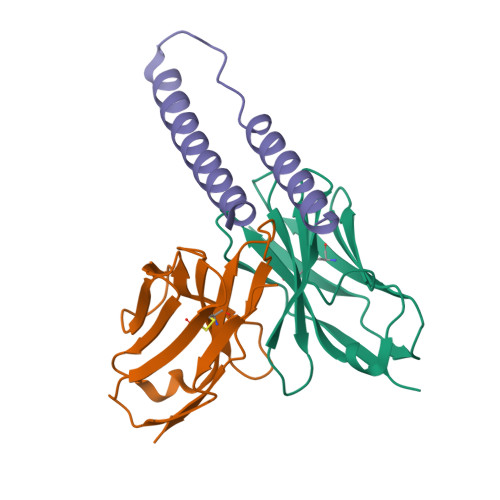
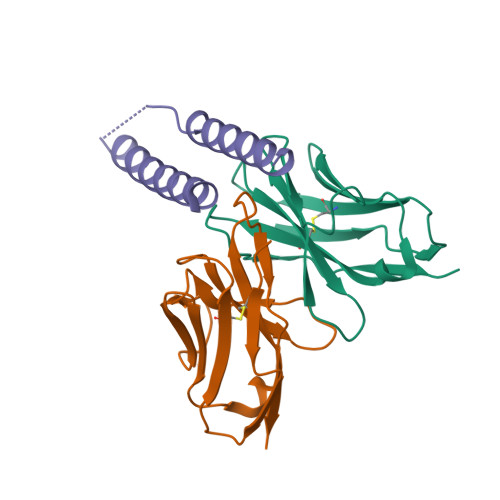
HIV-1 Rev NTD dimers with variable crossing anglesDiMattia, M.A., Watts, N.R., Wingfield, P.T., Grimes, J.M., Stuart, D.I., Steven, A.C. (2016) Structure 24: 1068-1080
HIV-1 Rev NTD dimers with variable crossing anglesDiMattia, M.A., Watts, N.R., Wingfield, P.T., Grimes, J.M., Stuart, D.I., Steven, A.C. (2016) Structure 24: 1068-1080
HIV-1 Rev NTD dimers with variable crossing anglesDiMattia, M.A., Watts, N.R., Wingfield, P.T., Grimes, J.M., Stuart, D.I., Steven, A.C. (2016) Structure 24: 1068-1080
HIV-1 Rev NTD dimers with variable crossing anglesDiMattia, M.A., Watts, N.R., Wingfield, P.T., Grimes, J.M., Stuart, D.I., Steven, A.C. (2016) Structure 24: 1068-1080
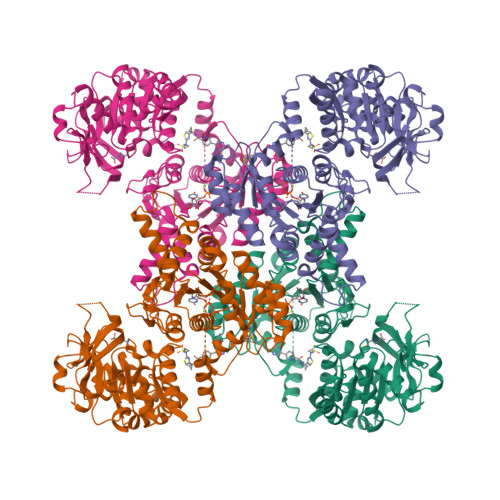
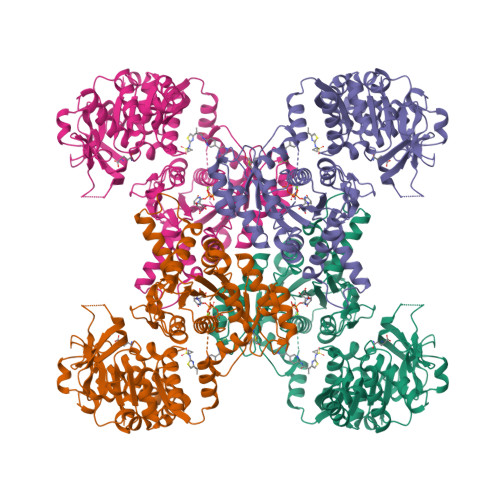
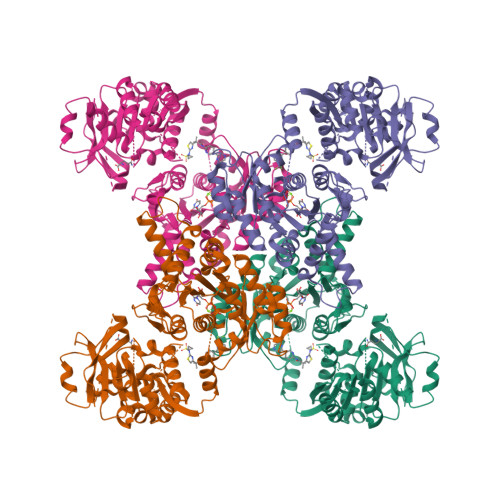
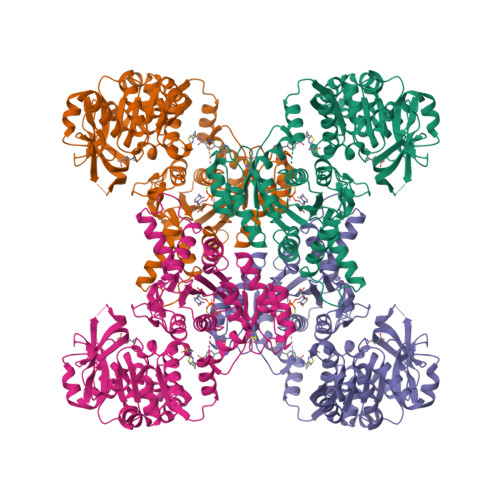
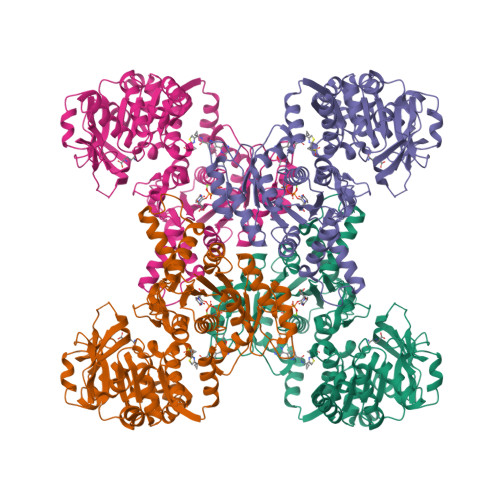
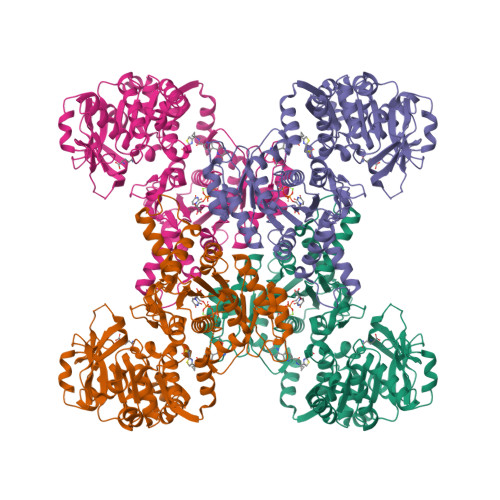
Human CTPS1 bound to UTP, AMPPNP, and glutamineLynch, E.M., Dimattia, M.A., Kollman, J.M. (2021) Proc Natl Acad Sci U S A 118:
Human CTPS1 bound to inhibitor R80Lynch, E.M., Dimattia, M.A., Kollman, J.M. (2021) Proc Natl Acad Sci U S A 118:
Human CTPS1 bound to inhibitor T35Lynch, E.M., Dimattia, M.A., Kollman, J.M. (2021) Proc Natl Acad Sci U S A 118:
Human CTPS2 bound to inhibitor R80Lynch, E.M., Dimattia, M.A., Kollman, J.M. (2021) Proc Natl Acad Sci U S A 118:
Human CTPS2 bound to inhibitor T35Lynch, E.M., Dimattia, M.A., Kollman, J.M. (2021) Proc Natl Acad Sci U S A 118:
Mouse CTPS1 bound to inhibitor R80Lynch, E.M., Dimattia, M.A., Kollman, J.M. (2021) Proc Natl Acad Sci U S A 118:
Mouse CTPS2 bound to inhibitor R80Lynch, E.M., Dimattia, M.A., Kollman, J.M. (2021) Proc Natl Acad Sci U S A 118:
Mouse CTPS2-I250T bound to inhibitor R80Lynch, E.M., Dimattia, M.A., Kollman, J.M. (2021) Proc Natl Acad Sci U S A 118:
Human RANBP2/RAN(GTP)/RANGAP1-SUMO1/UBC9/CRM1/RAN(GTP) - composite map and modelTo be published
1 to 17 of 17 Structures Page 1 of 1 Sort by |