Chimeric deubiquitinase engineering reveals structural basis for specific inhibition of the mitophagy regulator USP30
Kazi, N.H., Klink, N., Gallant, K., Kipka, G.M., Gersch, M.(2025) Nat Struct Mol Biol
Experimental Data Snapshot
Starting Models: in silico, experimental
View more details
(2025) Nat Struct Mol Biol
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ubiquitin carboxyl-terminal hydrolase 30,Ubiquitin carboxyl-terminal hydrolase 14,Ubiquitin carboxyl-terminal hydrolase 35 | 317 | Homo sapiens | Mutation(s): 1 Gene Names: USP30, USP14, TGT, USP35, KIAA1372, USP34 EC: 3.4.19.12 | 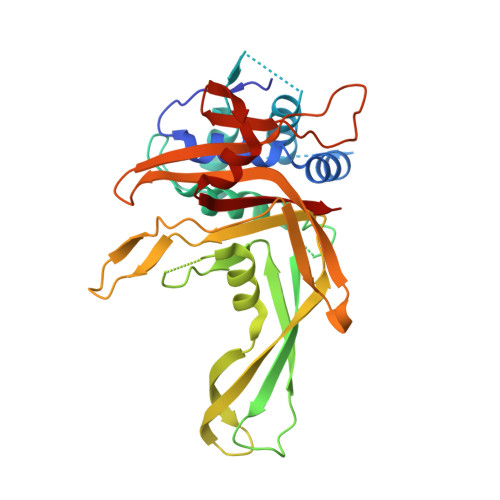 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P54578 (Homo sapiens) Explore P54578 Go to UniProtKB: P54578 | |||||
PHAROS: P54578 GTEx: ENSG00000101557 | |||||
Find proteins for Q9P2H5 (Homo sapiens) Explore Q9P2H5 Go to UniProtKB: Q9P2H5 | |||||
PHAROS: Q9P2H5 GTEx: ENSG00000118369 | |||||
Find proteins for Q70CQ3 (Homo sapiens) Explore Q70CQ3 Go to UniProtKB: Q70CQ3 | |||||
PHAROS: Q70CQ3 GTEx: ENSG00000135093 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | P54578Q70CQ3Q9P2H5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Polyubiquitin-B | 75 | Homo sapiens | Mutation(s): 0 Gene Names: UBB | 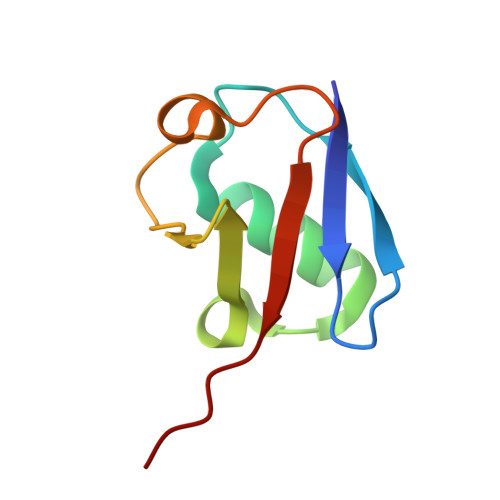 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P0CG47 (Homo sapiens) Explore P0CG47 Go to UniProtKB: P0CG47 | |||||
PHAROS: P0CG47 GTEx: ENSG00000170315 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0CG47 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AYE (Subject of Investigation/LOI) Query on AYE | C [auth A] | prop-2-en-1-amine C3 H7 N VVJKKWFAADXIJK-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 46.639 | α = 90 |
| b = 50.706 | β = 90 |
| c = 156.56 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| DIALS | data reduction |
| xia2 | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Coot | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Max Planck Society | Germany | CGCIII-352S |