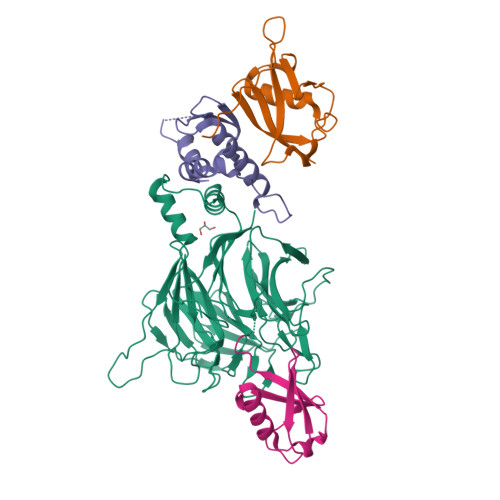
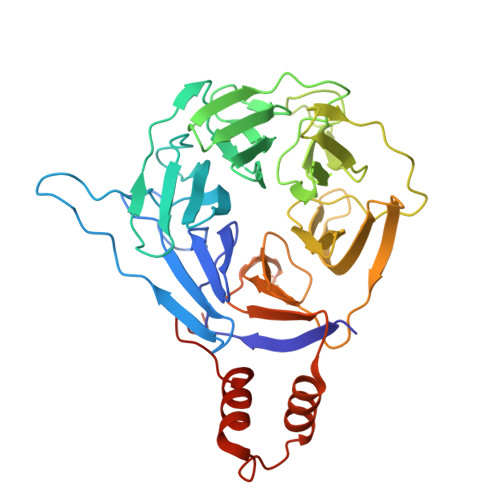
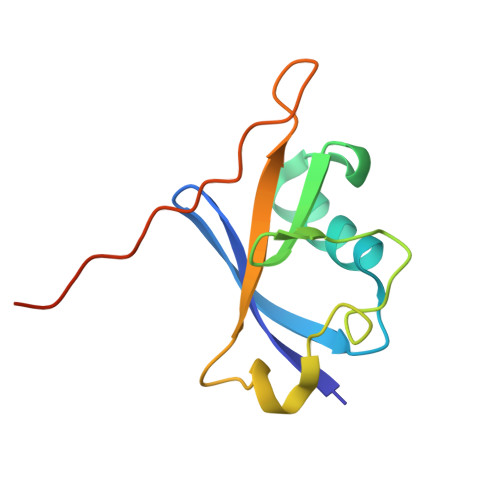
Structural basis for C-degron selectivity across KLHDCX family E3 ubiquitin ligases.
Scott, D.C., Chittori, S., Purser, N., King, M.T., Maiwald, S.A., Churion, K., Nourse, A., Lee, C., Paulo, J.A., Miller, D.J., Elledge, S.J., Harper, J.W., Kleiger, G., Schulman, B.A.(2024) Nat Commun 15: 9899-9899
- PubMed: 39548056
- DOI: https://doi.org/10.1038/s41467-024-54126-z
- Primary Citation of Related Structures:
9D1I, 9D1Y, 9D1Z, 9D8P - PubMed Abstract:
Specificity of the ubiquitin-proteasome system depends on E3 ligase-substrate interactions. Many such pairings depend on E3 ligases binding to peptide-like sequences - termed N- or C-degrons - at the termini of substrates. However, our knowledge of structural features distinguishing closely related C-degron substrate-E3 pairings is limited. Here, by systematically comparing ubiquitylation activities towards a suite of common model substrates, and defining interactions by biochemistry, crystallography, and cryo-EM, we reveal principles of C-degron recognition across the KLHDCX family of Cullin-RING ligases (CRLs). First, a motif common across these E3 ligases anchors a substrate's C-terminus. However, distinct locations of this C-terminus anchor motif in different blades of the KLHDC2, KLHDC3, and KLHDC10 β-propellers establishes distinct relative positioning and molecular environments for substrate C-termini. Second, our structural data show KLHDC3 has a pre-formed pocket establishing preference for an Arg or Gln preceding a C-terminal Gly, whereas conformational malleability contributes to KLHDC10's recognition of varying features adjacent to substrate C-termini. Finally, additional non-consensus interactions, mediated by C-degron binding grooves and/or by distal propeller surfaces and substrate globular domains, can substantially impact substrate binding and ubiquitylatability. Overall, the data reveal combinatorial mechanisms determining specificity and plasticity of substrate recognition by KLDCX-family C-degron E3 ligases.
Organizational Affiliation:
Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, TN, USA.