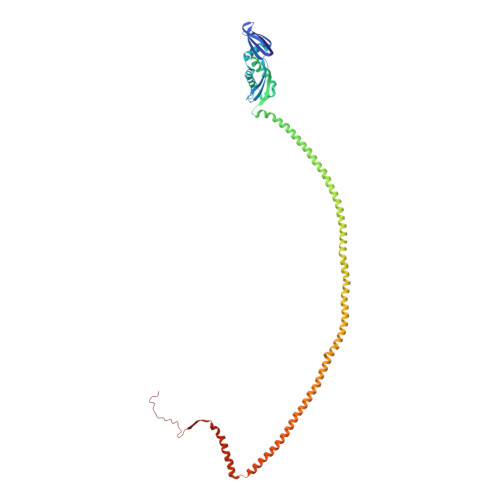
Structure of the flotillin complex in a native membrane environment.
Fu, Z., MacKinnon, R.(2024) Proc Natl Acad Sci U S A 121: e2409334121-e2409334121
- PubMed: 38985763
- DOI: https://doi.org/10.1073/pnas.2409334121
- Primary Citation of Related Structures:
9BQ2 - PubMed Abstract:
In this study, we used cryoelectron microscopy to determine the structures of the Flotillin protein complex, part of the Stomatin, Prohibitin, Flotillin, and HflK/C (SPFH) superfamily, from cell-derived vesicles without detergents. It forms a right-handed helical barrel consisting of 22 pairs of Flotillin1 and Flotillin2 subunits, with a diameter of 32 nm at its wider end and 19 nm at its narrower end. Oligomerization is stabilized by the C terminus, which forms two helical layers linked by a β-strand, and coiled-coil domains that enable strong charge-charge intersubunit interactions. Flotillin interacts with membranes at both ends; through its SPFH1 domains at the wide end and the C terminus at the narrow end, facilitated by hydrophobic interactions and lipidation. The inward tilting of the SPFH domain, likely triggered by phosphorylation, suggests its role in membrane curvature induction, which could be connected to its proposed role in clathrin-independent endocytosis. The structure suggests a shared architecture across the family of SPFH proteins and will promote further research into Flotillin's roles in cell biology.
Organizational Affiliation:
Laboratory of Molecular Neurobiology and Biophysics, The Rockefeller University, New York, NY 10065.