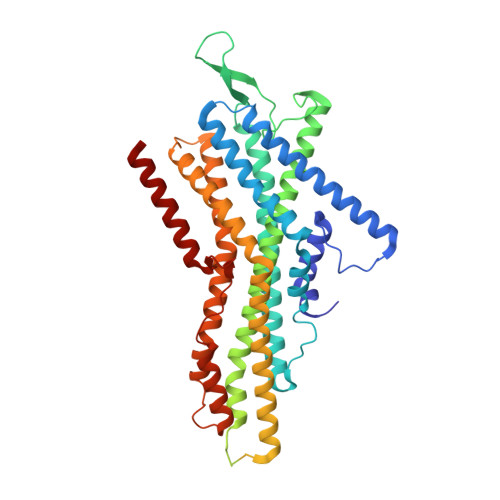
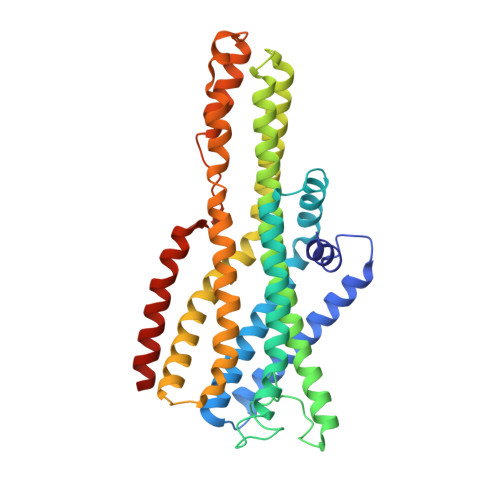
Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Wang, Y., Qiu, L., Wang, B., Guan, Z., Dong, Z., Zhang, J., Cao, S., Yang, L., Wang, B., Gong, Z., Zhang, L., Ma, W., Liu, Z., Zhang, D., Wang, G., Yin, P.(2024) Science 384: 1453-1460
- PubMed: 38870272
- DOI: https://doi.org/10.1126/science.adn6881
- Primary Citation of Related Structures:
8Z9A, 8Z9Z - PubMed Abstract:
Insects detect and discriminate a diverse array of chemicals using odorant receptors (ORs), which are ligand-gated ion channels comprising a divergent odorant-sensing OR and a conserved odorant receptor co-receptor (Orco). In this work, we report structures of the Ap OR5-Orco heterocomplex from the pea aphid Acyrthosiphon pisum alone and bound to its known activating ligand, geranyl acetate. In these structures, three Ap Orco subunits serve as scaffold components that cannot bind the ligand and remain relatively unchanged. Upon ligand binding, the pore-forming helix S7b of Ap OR5 shifts outward from the central pore axis, causing an asymmetrical pore opening for ion influx. Our study provides insights into odorant recognition and channel gating of the OR-Orco heterocomplex and offers structural resources to support development of innovative insecticides and repellents for pest control.
Organizational Affiliation:
Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Synthetic Biology Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518120, China.